N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization
- PMID: 17634239
- PMCID: PMC2045459
- DOI: 10.1128/JVI.01106-07
N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization
Abstract
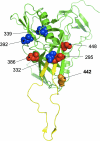
The monoclonal antibody (MAb) 2G12 recognizes a cluster of high-mannose oligosaccharides on the human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein gp120 and is one of a select group of MAbs with broad neutralizing activity. However, subtype C viruses are generally resistant to 2G12 neutralization. This has been attributed to the absence of a glycosylation site at position 295 in most subtype C gp120s, which instead is typically occupied by a Val residue. Here we show that N-linked glycans in addition to the one at position 295 are important in the formation of the 2G12 epitope in subtype C gp120. Introduction of the glycosylation site at position 295 into three subtype C molecular clones, Du151.2, COT9.6, and COT6.15, did increase 2G12 binding to all three mutagenized gp120s, but at various levels. The COT9-V295N mutant showed the strongest 2G12 binding and was the only mutant to become sensitive to 2G12 neutralization, although very high antibody concentrations were required. Introduction of a glycosylation site at position 448 into mutant COT6-V295N, which occurs naturally in COT9, resulted in a virus that was partially sensitive to 2G12. Interestingly, a glycosylation site at position 442, which is common among subtype C viruses, also contributed to the 2G12 epitope. The addition of this glycan increased virus neutralization sensitivity to 2G12, whereas its deletion conferred resistance. Collectively, our results indicate that the 2G12 binding site cannot readily be reconstituted on the envelopes of subtype C viruses, suggesting structural differences from other HIV subtypes in which the 2G12 epitope is naturally expressed.
Figures

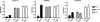
Similar articles
-
The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120.J Virol. 2002 Jul;76(14):7293-305. doi: 10.1128/jvi.76.14.7293-7305.2002. J Virol. 2002. PMID: 12072528 Free PMC article.
-
Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1.J Virol. 1996 Feb;70(2):1100-8. doi: 10.1128/JVI.70.2.1100-1108.1996. J Virol. 1996. PMID: 8551569 Free PMC article.
-
An engineered Saccharomyces cerevisiae strain binds the broadly neutralizing human immunodeficiency virus type 1 antibody 2G12 and elicits mannose-specific gp120-binding antibodies.J Virol. 2008 Jul;82(13):6447-57. doi: 10.1128/JVI.00412-08. Epub 2008 Apr 23. J Virol. 2008. PMID: 18434410 Free PMC article.
-
The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120.J Virol. 2002 Jul;76(14):7306-21. doi: 10.1128/jvi.76.14.7306-7321.2002. J Virol. 2002. PMID: 12072529 Free PMC article.
-
Antibody responses to the HIV-1 envelope high mannose patch.Adv Immunol. 2019;143:11-73. doi: 10.1016/bs.ai.2019.08.002. Epub 2019 Sep 11. Adv Immunol. 2019. PMID: 31607367 Free PMC article. Review.
Cited by
-
UCLA1, a synthetic derivative of a gp120 RNA aptamer, inhibits entry of human immunodeficiency virus type 1 subtype C.J Virol. 2012 May;86(9):4989-99. doi: 10.1128/JVI.06893-11. Epub 2012 Feb 29. J Virol. 2012. PMID: 22379083 Free PMC article.
-
HIV-1 gp120 determinants proximal to the CD4 binding site shift protective glycans that are targeted by monoclonal antibody 2G12.J Virol. 2010 Sep;84(18):9608-12. doi: 10.1128/JVI.00185-10. Epub 2010 Jul 7. J Virol. 2010. PMID: 20610714 Free PMC article.
-
Influences on the Design and Purification of Soluble, Recombinant Native-Like HIV-1 Envelope Glycoprotein Trimers.J Virol. 2015 Dec;89(23):12189-210. doi: 10.1128/JVI.01768-15. Epub 2015 Aug 26. J Virol. 2015. PMID: 26311893 Free PMC article.
-
Appreciating HIV type 1 diversity: subtype differences in Env.AIDS Res Hum Retroviruses. 2009 Mar;25(3):237-48. doi: 10.1089/aid.2008.0219. AIDS Res Hum Retroviruses. 2009. PMID: 19327047 Free PMC article.
-
Glycosylation site-specific analysis of clade C HIV-1 envelope proteins.J Proteome Res. 2009 Sep;8(9):4231-42. doi: 10.1021/pr9002728. J Proteome Res. 2009. PMID: 19610667 Free PMC article.
References
-
- Baba, T. W., V. Liska, R. Hofmann-Lehmann, J. Vlasak, W. Xu, S. Ayehunie, L. A. Cavacini, M. R. Posner, H. Katinger, G. Stiegler, B. J. Bernacky, T. A. Rizvi, R. Schmidt, L. R. Hill, M. E. Keeling, Y. Lu, J. E. Wright, T. C. Chou, and R. M. Ruprecht. 2000. Human neutralizing monoclonal antibodies of the IgG1 subtype protect against mucosal simian-human immunodeficiency virus infection. Nat. Med. 6:200-206. - PubMed
-
- Baribaud, F., S. Pohlmann, and R. W. Doms. 2001. The role of DC-SIGN and DC-SIGNR in HIV and SIV attachment, infection, and transmission. Virology 286:1-6. - PubMed
-
- Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. - PMC - PubMed
-
- Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

