A protein interaction map of the mitotic spindle
- PMID: 17634282
- PMCID: PMC1995735
- DOI: 10.1091/mbc.e07-06-0536
A protein interaction map of the mitotic spindle
Abstract
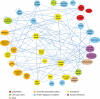
The mitotic spindle consists of a complex network of proteins that segregates chromosomes in eukaryotes. To strengthen our understanding of the molecular composition, organization, and regulation of the mitotic spindle, we performed a system-wide two-hybrid screen on 94 proteins implicated in spindle function in Saccharomyces cerevisiae. We report 604 predominantly novel interactions that were detected in multiple screens, involving 303 distinct prey proteins. We uncovered a pattern of extensive interactions between spindle proteins reflecting the intricate organization of the spindle. Furthermore, we observed novel connections between kinetochore complexes and chromatin-modifying proteins and used phosphorylation site mutants of NDC80/TID3 to gain insights into possible phospho-regulation mechanisms. We also present analyses of She1p, a novel spindle protein that interacts with the Dam1 kinetochore/spindle complex. The wealth of protein interactions presented here highlights the extent to which mitotic spindle protein functions and regulation are integrated with each other and with other cellular activities.
Figures


Similar articles
-
Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p.Mol Biol Cell. 2003 Aug;14(8):3342-55. doi: 10.1091/mbc.e02-11-0765. Epub 2003 May 3. Mol Biol Cell. 2003. PMID: 12925767 Free PMC article.
-
Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity.Mol Biol Cell. 2001 Sep;12(9):2601-13. doi: 10.1091/mbc.12.9.2601. Mol Biol Cell. 2001. PMID: 11553702 Free PMC article.
-
Molecular architecture and assembly of the yeast kinetochore MIND complex.J Cell Biol. 2010 Sep 6;190(5):823-34. doi: 10.1083/jcb.201002059. J Cell Biol. 2010. PMID: 20819936 Free PMC article.
-
Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans.Trends Cell Biol. 2013 Jun;23(6):260-9. doi: 10.1016/j.tcb.2013.01.010. Epub 2013 Mar 5. Trends Cell Biol. 2013. PMID: 23481674 Review.
-
Coupling spindle position with mitotic exit in budding yeast: The multifaceted role of the small GTPase Tem1.Small GTPases. 2015 Oct 2;6(4):196-201. doi: 10.1080/21541248.2015.1109023. Small GTPases. 2015. PMID: 26507466 Free PMC article. Review.
Cited by
-
Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases.Genes (Basel). 2020 Feb 18;11(2):205. doi: 10.3390/genes11020205. Genes (Basel). 2020. PMID: 32085395 Free PMC article. Review.
-
Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches.PLoS Genet. 2011 Mar;7(3):e1002008. doi: 10.1371/journal.pgen.1002008. Epub 2011 Mar 3. PLoS Genet. 2011. PMID: 21408204 Free PMC article.
-
Microtubule cross-linking activity of She1 ensures spindle stability for spindle positioning.J Cell Biol. 2017 Sep 4;216(9):2759-2775. doi: 10.1083/jcb.201701094. Epub 2017 Aug 9. J Cell Biol. 2017. PMID: 28794129 Free PMC article.
-
The yeast homologue of the microtubule-associated protein Lis1 interacts with the sumoylation machinery and a SUMO-targeted ubiquitin ligase.Mol Biol Cell. 2012 Dec;23(23):4552-66. doi: 10.1091/mbc.E12-03-0195. Epub 2012 Oct 3. Mol Biol Cell. 2012. PMID: 23034179 Free PMC article.
-
Histone H3K4 methylation regulates deactivation of the spindle assembly checkpoint through direct binding of Mad2.Genes Dev. 2016 May 15;30(10):1187-97. doi: 10.1101/gad.278887.116. Epub 2016 May 19. Genes Dev. 2016. PMID: 27198228 Free PMC article.
References
-
- Ayscough K. R., Drubin D. G. Cell Biology: A Laboratory Handbook. Vol. 2. San Diego: Academic Press; 1998. Immunofluorescence microscopy of yeast cells; pp. 477–485.
-
- Biddick R., Young E. T. Yeast mediator and its role in transcriptional regulation. CR Biol. 2005;328:773–782. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

