Genomewide association analysis of coronary artery disease
- PMID: 17634449
- PMCID: PMC2719290
- DOI: 10.1056/NEJMoa072366
Genomewide association analysis of coronary artery disease
Abstract
Background: Modern genotyping platforms permit a systematic search for inherited components of complex diseases. We performed a joint analysis of two genomewide association studies of coronary artery disease.
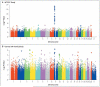
Methods: We first identified chromosomal loci that were strongly associated with coronary artery disease in the Wellcome Trust Case Control Consortium (WTCCC) study (which involved 1926 case subjects with coronary artery disease and 2938 controls) and looked for replication in the German MI [Myocardial Infarction] Family Study (which involved 875 case subjects with myocardial infarction and 1644 controls). Data on other single-nucleotide polymorphisms (SNPs) that were significantly associated with coronary artery disease in either study (P<0.001) were then combined to identify additional loci with a high probability of true association. Genotyping in both studies was performed with the use of the GeneChip Human Mapping 500K Array Set (Affymetrix).
Results: Of thousands of chromosomal loci studied, the same locus had the strongest association with coronary artery disease in both the WTCCC and the German studies: chromosome 9p21.3 (SNP, rs1333049) (P=1.80x10(-14) and P=3.40x10(-6), respectively). Overall, the WTCCC study revealed nine loci that were strongly associated with coronary artery disease (P<1.2x10(-5) and less than a 50% chance of being falsely positive). In addition to chromosome 9p21.3, two of these loci were successfully replicated (adjusted P<0.05) in the German study: chromosome 6q25.1 (rs6922269) and chromosome 2q36.3 (rs2943634). The combined analysis of the two studies identified four additional loci significantly associated with coronary artery disease (P<1.3x10(-6)) and a high probability (>80%) of a true association: chromosomes 1p13.3 (rs599839), 1q41 (rs17465637), 10q11.21 (rs501120), and 15q22.33 (rs17228212).
Conclusions: We identified several genetic loci that, individually and in aggregate, substantially affect the risk of development of coronary artery disease.
Copyright 2007 Massachusetts Medical Society.
Figures
Comment in
-
Drinking from the fire hose--statistical issues in genomewide association studies.N Engl J Med. 2007 Aug 2;357(5):436-9. doi: 10.1056/NEJMp078120. Epub 2007 Jul 18. N Engl J Med. 2007. PMID: 17634446 No abstract available.
-
Scanning the genome for coronary risk.N Engl J Med. 2007 Aug 2;357(5):497-9. doi: 10.1056/NEJMe078121. Epub 2007 Jul 18. N Engl J Med. 2007. PMID: 17634453 No abstract available.
References
-
- Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367:1747–57. - PubMed
-
- Yusuf S, Hawken S, Ounpuu S, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364:937–52. - PubMed
-
- Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–6. - PubMed
-
- Christensen K, Murray JC. What genome-wide association studies can do for medicine. N Engl J Med. 2007;356:1094–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
