Human nuclear pregnane X receptor cross-talk with CREB to repress cAMP activation of the glucose-6-phosphatase gene
- PMID: 17635106
- PMCID: PMC2275060
- DOI: 10.1042/BJ20070481
Human nuclear pregnane X receptor cross-talk with CREB to repress cAMP activation of the glucose-6-phosphatase gene
Abstract
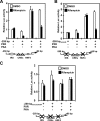
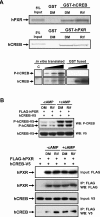
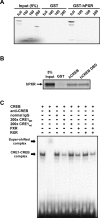
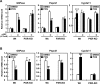
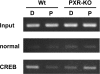
The nuclear PXR (pregnane X receptor) was originally characterized as a key transcription factor that activated hepatic genes encoding drug-metabolizing enzymes. We have now demonstrated that PXR also represses glucagon-activated transcription of the G6Pase (glucose-6-phosphatase) gene by directly binding to CREB [CRE (cAMP-response element)-binding protein]. Adenoviral-mediated expression of human PXR (hPXR) and its activation by rifampicin strongly repressed cAMP-dependent induction of the endogenous G6Pase gene in Huh7 cells. Using the -259 bp G6Pase promoter construct in cell-based transcription assays, repression by hPXR of PKA (cAMP-dependent protein kinase)-mediated promoter activation was delineated to CRE sites. GST (glutathione transferase) pull-down and immunoprecipitation assays were employed to show that PXR binds directly to CREB, while gel-shift assays were used to demonstrate that this binding prevents CREB interaction with the CRE. These results are consistent with the hypothesis that PXR represses the transcription of the G6Pase gene by inhibiting the DNA-binding ability of CREB. In support of this hypothesis, treatment with the mouse PXR activator PCN (pregnenolone 16alpha-carbonitrile) repressed cAMP-dependent induction of the G6Pase gene in primary hepatocytes prepared from wild-type, but not from PXR-knockout, mice, and also in the liver of fasting wild-type, but not PXR-knockout, mice. Moreover, ChIP (chromatin immunoprecipitation) assays were performed to show a decreased CREB binding to the G6Pase promoter in fasting wild-type mice after PCN treatment. Thus drug activation of PXR can repress the transcriptional activity of CREB, down-regulating gluconeogenesis.
Figures



Similar articles
-
Nuclear pregnane X receptor cross-talk with FoxA2 to mediate drug-induced regulation of lipid metabolism in fasting mouse liver.J Biol Chem. 2007 Mar 30;282(13):9768-9776. doi: 10.1074/jbc.M610072200. Epub 2007 Jan 30. J Biol Chem. 2007. PMID: 17267396 Free PMC article.
-
Nuclear receptors CAR and PXR cross talk with FOXO1 to regulate genes that encode drug-metabolizing and gluconeogenic enzymes.Mol Cell Biol. 2004 Sep;24(18):7931-40. doi: 10.1128/MCB.24.18.7931-7940.2004. Mol Cell Biol. 2004. PMID: 15340055 Free PMC article.
-
Crosstalk of CREB and Fos/Jun on a single cis-element: transcriptional repression of the steroidogenic acute regulatory protein gene.J Mol Endocrinol. 2007 Oct;39(4):261-77. doi: 10.1677/JME-07-0065. J Mol Endocrinol. 2007. PMID: 17909266
-
Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM.Exp Cell Res. 2002 May 1;275(2):143-54. doi: 10.1006/excr.2002.5491. Exp Cell Res. 2002. PMID: 11969286 Review.
-
The roles of nuclear receptors CAR and PXR in hepatic energy metabolism.Drug Metab Pharmacokinet. 2008;23(1):8-13. doi: 10.2133/dmpk.23.8. Drug Metab Pharmacokinet. 2008. PMID: 18305370 Review.
Cited by
-
Pregnenolone 16-Alpha Carbonitrile, an Agonist of Rodent Pregnane X Receptor, Regulates Testosterone Biosynthesis in Rodent Leydig Cells.J Xenobiot. 2024 Sep 16;14(3):1256-1267. doi: 10.3390/jox14030071. J Xenobiot. 2024. PMID: 39311150 Free PMC article.
-
A selective gut bacterial bile salt hydrolase alters host metabolism.Elife. 2018 Jul 17;7:e37182. doi: 10.7554/eLife.37182. Elife. 2018. PMID: 30014852 Free PMC article.
-
Liganded pregnane X receptor represses the human sulfotransferase SULT1E1 promoter through disrupting its chromatin structure.Nucleic Acids Res. 2011 Oct;39(19):8392-403. doi: 10.1093/nar/gkr458. Epub 2011 Jul 14. Nucleic Acids Res. 2011. PMID: 21764778 Free PMC article.
-
The Three Ds of Transcription Activation by Glucagon: Direct, Delayed, and Dynamic.Endocrinology. 2018 Jan 1;159(1):206-216. doi: 10.1210/en.2017-00521. Endocrinology. 2018. PMID: 29077799 Free PMC article. Review.
-
PXR and 4β-Hydroxycholesterol Axis and the Components of Metabolic Syndrome.Cells. 2020 Nov 9;9(11):2445. doi: 10.3390/cells9112445. Cells. 2020. PMID: 33182477 Free PMC article. Review.
References
-
- Hanson R. W., Reshef L. Regulation of phosphoenolpyruvate carboxykinase (GTP) gene expression. Annu. Rev. Biochem. 1997;66:581–611. - PubMed
-
- Schmoll D., Wasner C., Hinds C. J., Allan B. B., Walther R., Burchell A. Identification of a cAMP response element within the glucose-6-phosphatase hydrolytic subunit gene promoter which is involved in the transcriptional regulation by cAMP and glucocorticoids in H4IIE hepatoma cells. Biochem. J. 1999;338:457–463. - PMC - PubMed
-
- Yoon J. C., Puigserver P., Chen G., Donovan J., Wu Z., Rhee J., Adelmant G., Stafford J., Kahn C. R., Granner D. K., et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

