Activation of human CD4+ T cells by targeting MHC class II epitopes to endosomal compartments using human CD1 tail sequences
- PMID: 17635609
- PMCID: PMC2266034
- DOI: 10.1111/j.1365-2567.2007.02666.x
Activation of human CD4+ T cells by targeting MHC class II epitopes to endosomal compartments using human CD1 tail sequences
Abstract
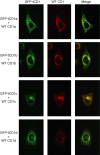
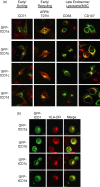
Distinct CD4(+) T-cell epitopes within the same protein can be optimally processed and loaded into major histocompatibility complex (MHC) class II molecules in disparate endosomal compartments. The CD1 protein isoforms traffic to these same endosomal compartments as directed by unique cytoplasmic tail sequences, therefore we reasoned that antigen/CD1 chimeras containing the different CD1 cytoplasmic tail sequences could optimally target antigens to the MHC class II antigen presentation pathway. Evaluation of trafficking patterns revealed that all four human CD1-derived targeting sequences delivered antigen to the MHC class II antigen presentation pathway, to early/recycling, early/sorting and late endosomes/lysosomes. There was a preferential requirement for different CD1 targeting sequences for the optimal presentation of an MHC class II epitope in the following hierarchy: CD1b > CD1d = CD1c > > > CD1a or untargeted antigen. Therefore, the substitution of the CD1 ectodomain with heterologous proteins results in their traffic to distinct intracellular locations that intersect with MHC class II and this differential distribution leads to specific functional outcomes with respect to MHC class II antigen presentation. These findings may have implications in designing DNA vaccines, providing a greater variety of tools to generate T-cell responses against microbial pathogens or tumours.
Figures


Similar articles
-
CD1 and MHC II find different means to the same end.Trends Immunol. 2005 May;26(5):282-8. doi: 10.1016/j.it.2005.03.002. Trends Immunol. 2005. PMID: 15866242 Review.
-
Melanosomal targeting sequences from gp100 are essential for MHC class II-restricted endogenous epitope presentation and mobilization to endosomal compartments.Cancer Res. 2006 Feb 15;66(4):2423-32. doi: 10.1158/0008-5472.CAN-05-2516. Cancer Res. 2006. PMID: 16489049
-
The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules.Nat Immunol. 2004 Jul;5(7):685-92. doi: 10.1038/ni1088. Nat Immunol. 2004. PMID: 15224094 Review.
-
Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens.J Immunol. 2010 Feb 1;184(3):1235-41. doi: 10.4049/jimmunol.0804140. Epub 2009 Dec 21. J Immunol. 2010. PMID: 20026739
-
CD1--a new paradigm for antigen presentation and T cell activation.Clin Immunol Immunopathol. 1998 Apr;87(1):8-14. doi: 10.1006/clin.1997.4500. Clin Immunol Immunopathol. 1998. PMID: 9576005 Review.
Cited by
-
Mucosal COVID-19 vaccines: Risks, benefits and control of the pandemic.World J Virol. 2022 Sep 25;11(5):221-236. doi: 10.5501/wjv.v11.i5.221. World J Virol. 2022. PMID: 36188733 Free PMC article. Review.
-
Conserved mycobacterial lipoglycoproteins activate TLR2 but also require glycosylation for MHC class II-restricted T cell activation.J Immunol. 2008 May 1;180(9):5833-42. doi: 10.4049/jimmunol.180.9.5833. J Immunol. 2008. PMID: 18424702 Free PMC article.
-
Interleukin-1β triggers the differentiation of macrophages with enhanced capacity to present mycobacterial antigen to T cells.Immunology. 2014 Feb;141(2):174-80. doi: 10.1111/imm.12167. Immunology. 2014. PMID: 24032597 Free PMC article.
-
The future of human DNA vaccines.J Biotechnol. 2012 Dec 31;162(2-3):171-82. doi: 10.1016/j.jbiotec.2012.08.012. Epub 2012 Sep 7. J Biotechnol. 2012. PMID: 22981627 Free PMC article. Review.
-
Dual-Antigen COVID-19 Vaccine Subcutaneous Prime Delivery With Oral Boosts Protects NHP Against SARS-CoV-2 Challenge.Front Immunol. 2021 Sep 16;12:729837. doi: 10.3389/fimmu.2021.729837. eCollection 2021. Front Immunol. 2021. PMID: 34603305 Free PMC article.
References
-
- Trombetta ES, Mellman I. Cell biology of antigen processing in vitro and in vivo. Annu Rev Immunol. 2005;23:975–1028. - PubMed
-
- Honey K, Rudensky AY. Lysosomal cysteine proteases regulate antigen presentation. Nat Rev Immunol. 2003;3:472–82. - PubMed
-
- Watts C. Capture and processing of exogenous antigens for presentation on MHC molecules. Annu Rev Immunol. 1997;15:821–50. - PubMed
-
- Pathak SS, Blum JS. Endocytic recycling is required for the presentation of an exogenous peptide via MHC class II molecules. Traffic. 2000;1:561–9. - PubMed
-
- Delvig AA, Robinson JH. Different endosomal proteolysis requirements for antigen processing of two T-cell epitopes of the M5 protein from viable Streptococcus pyogenes. J Biol Chem. 1998;273:3291–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

