Molecular characterization of the Aedes aegypti odorant receptor gene family
- PMID: 17635615
- PMCID: PMC3100214
- DOI: 10.1111/j.1365-2583.2007.00748.x
Molecular characterization of the Aedes aegypti odorant receptor gene family
Abstract
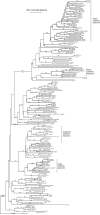
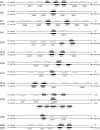
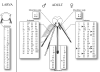
The olfactory-driven blood-feeding behaviour of female Aedes aegypti mosquitoes is the primary transmission mechanism by which the arboviruses causing dengue and yellow fevers affect over 40 million individuals worldwide. Bioinformatics analysis has been used to identify 131 putative odourant receptors from the A. aegypti genome that are likely to function in chemosensory perception in this mosquito. Comparison with the Anopheles gambiae olfactory subgenome demonstrates significant divergence of the odourant receptors that reflects a high degree of evolutionary activity potentially resulting from their critical roles during the mosquito life cycle. Expression analyses in the larval and adult olfactory chemosensory organs reveal that the ratio of odourant receptors to antennal glomeruli is not necessarily one to one in mosquitoes.
Figures

References
-
- Ache BW, Young JM. Olfaction: diverse species, conserved principles. Neuron. 2005;48:417–430. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Berger J, Suzuki T, Senti KA, Stubbs J, Schaffner G, Dickson BJ. Genetic mapping with snp markers in Drosophila. Nat Genet. 2001;29:475–481. - PubMed
-
- Buck L, Axel R. A novel mulitgene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

