Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals
- PMID: 17635934
- PMCID: PMC2064441
- DOI: 10.1083/jcb.200612031
Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals
Abstract
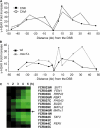
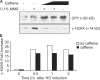
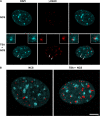
Double-strand break (DSB) damage in yeast and mammalian cells induces the rapid ATM (ataxia telangiectasia mutated)/ATR (ataxia telangiectasia and Rad3 related)-dependent phosphorylation of histone H2AX (gamma-H2AX). In budding yeast, a single endonuclease-induced DSB triggers gamma-H2AX modification of 50 kb on either side of the DSB. The extent of gamma-H2AX spreading does not depend on the chromosomal sequences. DNA resection after DSB formation causes the slow, progressive loss of gamma-H2AX from single-stranded DNA and, after several hours, the Mec1 (ATR)-dependent spreading of gamma-H2AX to more distant regions. Heterochromatic sequences are only weakly modified upon insertion of a 3-kb silent HMR locus into a gamma-H2AX-covered region. The presence of heterochromatin does not stop the phosphorylation of chromatin more distant from the DSB. In mouse embryo fibroblasts, gamma-H2AX distribution shows that gamma-H2AX foci increase in size as chromatin becomes more accessible. In yeast, we see a high level of constitutive gamma-H2AX in telomere regions in the absence of any exogenous DNA damage, suggesting that yeast chromosome ends are transiently detected as DSBs.
Figures


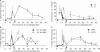

References
-
- Bloom, K.S., and J. Carbon. 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 29:305–317. - PubMed
-
- Celeste, A., O. Fernandez-Capetillo, M.J. Kruhlak, D.R. Pilch, D.W. Staudt, A. Lee, R.F. Bonner, W.M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

