Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4
- PMID: 17636017
- PMCID: PMC2099224
- DOI: 10.1128/MCB.00853-07
Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4
Abstract
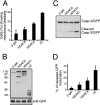
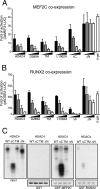
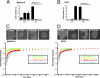
From the nucleus, histone deacetylase 4 (HDAC4) regulates a variety of cellular processes, including growth, differentiation, and survival, by orchestrating transcriptional changes. Extracellular signals control its repressive influence mostly through regulating its nuclear-cytoplasmic shuttling. In particular, specific posttranslational modifications such as phosphorylation and caspase-mediated proteolytic processing operate on HDAC4 to promote its nuclear accumulation or export. To understand the signaling properties of this deacetylase, we investigated its cell death-promoting activity and the transcriptional repression potential of different mutants that accumulate in the nucleus. Here we show that, compared to that of other nuclear forms of HDAC4, a caspase-generated nuclear fragment exhibits a stronger cell death-promoting activity coupled with increased repressive effect on Runx2- or SRF-dependent transcription. However, this mutant displays reduced repressive action on MEF2C-driven transcription. Photobleaching experiments and quantitative analysis of the raw data, based on a two-binding-state compartmental model, demonstrate the existence of two nuclear pools of HDAC4 with different chromatin-binding properties. The caspase-generated fragment is weakly bound to chromatin, whereas an HDAC4 mutant defective in 14-3-3 binding or the wild-type HDAC5 protein forms a more stable complex. The tightly bound species show an impaired ability to induce cell death and repress Runx2- or SRF-dependent transcription less efficiently. We propose that, through specific posttranslation modifications, extracellular signals control two distinct nuclear pools of HDAC4 to differentially dictate cell death and differentiation. These two nuclear pools of HDAC4 are characterized by different repression potentials and divergent dynamics of chromatin interaction.
Figures





Similar articles
-
Caspase-dependent regulation of histone deacetylase 4 nuclear-cytoplasmic shuttling promotes apoptosis.Mol Biol Cell. 2004 Jun;15(6):2804-18. doi: 10.1091/mbc.e03-08-0624. Epub 2004 Apr 9. Mol Biol Cell. 2004. PMID: 15075374 Free PMC article.
-
Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors.Mol Cell Biol. 2005 Mar;25(6):2273-87. doi: 10.1128/MCB.25.6.2273-2287.2005. Mol Cell Biol. 2005. PMID: 15743823 Free PMC article.
-
CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy.J Clin Invest. 2006 Jul;116(7):1853-64. doi: 10.1172/JCI27438. Epub 2006 Jun 8. J Clin Invest. 2006. PMID: 16767219 Free PMC article.
-
Beside the MEF2 axis: unconventional functions of HDAC4.Cell Signal. 2013 Jan;25(1):269-76. doi: 10.1016/j.cellsig.2012.10.002. Epub 2012 Oct 11. Cell Signal. 2013. PMID: 23063464 Review.
-
Class II histone deacetylases: structure, function, and regulation.Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review.
Cited by
-
The functional interactome landscape of the human histone deacetylase family.Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. Mol Syst Biol. 2013. PMID: 23752268 Free PMC article.
-
Subcellular relocation of histone deacetylase 4 regulates growth plate chondrocyte differentiation through Ca2+/calmodulin-dependent kinase IV.Am J Physiol Cell Physiol. 2012 Jul 1;303(1):C33-40. doi: 10.1152/ajpcell.00348.2011. Epub 2012 Mar 21. Am J Physiol Cell Physiol. 2012. PMID: 22442139 Free PMC article.
-
Histone deacetylase signaling in cardioprotection.Cell Mol Life Sci. 2014 May;71(9):1673-90. doi: 10.1007/s00018-013-1516-9. Epub 2013 Dec 6. Cell Mol Life Sci. 2014. PMID: 24310814 Free PMC article. Review.
-
Roles of Histone Deacetylase 4 in the Inflammatory and Metabolic Processes.Diabetes Metab J. 2024 May;48(3):340-353. doi: 10.4093/dmj.2023.0174. Epub 2024 Mar 22. Diabetes Metab J. 2024. PMID: 38514922 Free PMC article. Review.
-
Ubiquitin-dependent degradation of HDAC4, a new regulator of random cell motility.Mol Biol Cell. 2011 Jan 15;22(2):278-89. doi: 10.1091/mbc.E10-07-0616. Epub 2010 Nov 30. Mol Biol Cell. 2011. PMID: 21118993 Free PMC article.
References
-
- Bellido, T., A. A. Ali, L. I. Plotkin, Q. Fu, I. Gubrij, P. K. Roberson, R. S. Weinstein, C. A. O'Brien, S. C. Manolagas, and R. L. Jilka. 2003. Proteasomal degradation of Runx2 shortens parathyroid hormone-induced anti-apoptotic signaling in osteoblasts. A putative explanation for why intermittent administration is needed for bone anabolism. J. Biol. Chem. 278:50259-50272. - PubMed
-
- Blyth, K., F. Vaillant, L. Hanlon, N. Mackay, M. Bell, A. Jenkins, J. C. Neil, and E. R. Cameron. 2006. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 66:2195-2201. - PubMed
-
- Borghi, S., S. Molinari, G. Razzini, F. Parise, R. Battini, and S. Ferrari. 2001. The nuclear localization domain of the MEF2 family of transcription factors shows member-specific features and mediates the nuclear import of histone deacetylase 4. J. Cell Sci. 114:4477-4483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
