Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4
- PMID: 17636017
- PMCID: PMC2099224
- DOI: 10.1128/MCB.00853-07
Dephosphorylation and caspase processing generate distinct nuclear pools of histone deacetylase 4
Abstract
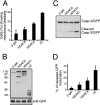
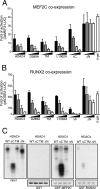
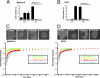
From the nucleus, histone deacetylase 4 (HDAC4) regulates a variety of cellular processes, including growth, differentiation, and survival, by orchestrating transcriptional changes. Extracellular signals control its repressive influence mostly through regulating its nuclear-cytoplasmic shuttling. In particular, specific posttranslational modifications such as phosphorylation and caspase-mediated proteolytic processing operate on HDAC4 to promote its nuclear accumulation or export. To understand the signaling properties of this deacetylase, we investigated its cell death-promoting activity and the transcriptional repression potential of different mutants that accumulate in the nucleus. Here we show that, compared to that of other nuclear forms of HDAC4, a caspase-generated nuclear fragment exhibits a stronger cell death-promoting activity coupled with increased repressive effect on Runx2- or SRF-dependent transcription. However, this mutant displays reduced repressive action on MEF2C-driven transcription. Photobleaching experiments and quantitative analysis of the raw data, based on a two-binding-state compartmental model, demonstrate the existence of two nuclear pools of HDAC4 with different chromatin-binding properties. The caspase-generated fragment is weakly bound to chromatin, whereas an HDAC4 mutant defective in 14-3-3 binding or the wild-type HDAC5 protein forms a more stable complex. The tightly bound species show an impaired ability to induce cell death and repress Runx2- or SRF-dependent transcription less efficiently. We propose that, through specific posttranslation modifications, extracellular signals control two distinct nuclear pools of HDAC4 to differentially dictate cell death and differentiation. These two nuclear pools of HDAC4 are characterized by different repression potentials and divergent dynamics of chromatin interaction.
Figures





References
-
- Bellido, T., A. A. Ali, L. I. Plotkin, Q. Fu, I. Gubrij, P. K. Roberson, R. S. Weinstein, C. A. O'Brien, S. C. Manolagas, and R. L. Jilka. 2003. Proteasomal degradation of Runx2 shortens parathyroid hormone-induced anti-apoptotic signaling in osteoblasts. A putative explanation for why intermittent administration is needed for bone anabolism. J. Biol. Chem. 278:50259-50272. - PubMed
-
- Blyth, K., F. Vaillant, L. Hanlon, N. Mackay, M. Bell, A. Jenkins, J. C. Neil, and E. R. Cameron. 2006. Runx2 and MYC collaborate in lymphoma development by suppressing apoptotic and growth arrest pathways in vivo. Cancer Res. 66:2195-2201. - PubMed
-
- Borghi, S., S. Molinari, G. Razzini, F. Parise, R. Battini, and S. Ferrari. 2001. The nuclear localization domain of the MEF2 family of transcription factors shows member-specific features and mediates the nuclear import of histone deacetylase 4. J. Cell Sci. 114:4477-4483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
