Replication in hydroxyurea: it's a matter of time
- PMID: 17636020
- PMCID: PMC2099622
- DOI: 10.1128/MCB.00719-07
Replication in hydroxyurea: it's a matter of time
Abstract
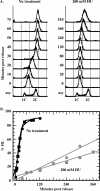
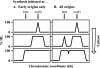
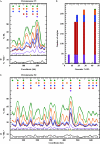
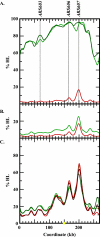
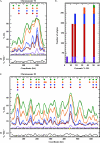
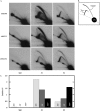
Hydroxyurea (HU) is a DNA replication inhibitor that negatively affects both the elongation and initiation phases of replication and triggers the "intra-S phase checkpoint." Previous work with budding yeast has shown that, during a short exposure to HU, MEC1/RAD53 prevent initiation at some late S phase origins. In this study, we have performed microarray experiments to follow the fate of all origins over an extended exposure to HU. We show that the genome-wide progression of DNA synthesis, including origin activation, follows the same pattern in the presence of HU as in its absence, although the time frames are very different. We find no evidence for a specific effect that excludes initiation from late origins. Rather, HU causes S phase to proceed in slow motion; all temporal classes of origins are affected, but the order in which they become active is maintained. We propose a revised model for the checkpoint response to HU that accounts for the continued but slowed pace of the temporal program of origin activation.
Figures
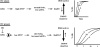
References
-
- Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. - PubMed
-
- Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. - PubMed
-
- Aparicio, O. M., D. M. Weinstein, and S. P. Bell. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91:59-69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
