An interaction between two RNA binding proteins, Nab2 and Pub1, links mRNA processing/export and mRNA stability
- PMID: 17636033
- PMCID: PMC2099604
- DOI: 10.1128/MCB.00881-07
An interaction between two RNA binding proteins, Nab2 and Pub1, links mRNA processing/export and mRNA stability
Abstract
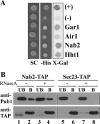
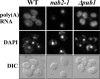
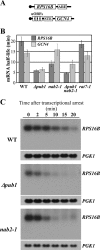
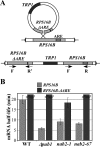
mRNA stability is modulated by elements in the mRNA transcript and their cognate RNA binding proteins. Poly(U) binding protein 1 (Pub1) is a cytoplasmic Saccharomyces cerevisiae mRNA binding protein that stabilizes transcripts containing AU-rich elements (AREs) or stabilizer elements (STEs). In a yeast two-hybrid screen, we identified nuclear poly(A) binding protein 2 (Nab2) as being a Pub1-interacting protein. Nab2 is an essential nucleocytoplasmic shuttling mRNA binding protein that regulates poly(A) tail length and mRNA export. The interaction between Pub1 and Nab2 was confirmed by copurification and in vitro binding assays. The interaction is mediated by the Nab2 zinc finger domain. Analysis of the functional link between these proteins reveals that Nab2, like Pub1, can modulate the stability of specific mRNA transcripts. The half-life of the RPS16B transcript, an ARE-like sequence-containing Pub1 target, is decreased in both nab2-1 and nab2-67 mutants. In contrast, GCN4, an STE-containing Pub1 target, is not affected. Similar results were obtained for other ARE- and STE-containing Pub1 target transcripts. Further analysis reveals that the ARE-like sequence is necessary for Nab2-mediated transcript stabilization. These results suggest that Nab2 functions together with Pub1 to modulate mRNA stability and strengthen a model where nuclear events are coupled to the control of mRNA turnover in the cytoplasm.
Figures


References
-
- Amberg, D. C., A. L. Goldstein, and C. N. Cole. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6:1173-1189. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
