The dawn of dominance by the mature domain in tRNA splicing
- PMID: 17636125
- PMCID: PMC1941465
- DOI: 10.1073/pnas.0705537104
The dawn of dominance by the mature domain in tRNA splicing
Abstract
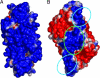
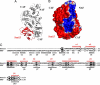
The relationship between enzyme architecture and substrate specificity among archaeal pre-tRNA splicing endonucleases has been investigated more deeply, by using biochemical assays and model building. The enzyme from Archeoglobus fulgidus (AF) is particularly interesting: it cleaves the bulge-helix-bulge target without requiring the mature tRNA domain, but, when the target is a bulge-helix-loop, the mature domain is required. A model of AF based on its electrostatic potential shows three polar patches interacting with the pre-tRNA substrate. A simple deletion mutant of the AF endonuclease lacking two of the three polar patches no longer cleaves the bulge-helix-loop substrate with or without the mature domain. This single deletion shows a possible path for the evolution of eukaryal splicing endonucleases from the archaeal enzyme.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
RNA recognition and cleavage by a splicing endonuclease.Science. 2006 May 12;312(5775):906-10. doi: 10.1126/science.1126629. Science. 2006. PMID: 16690865
-
Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus.J Mol Biol. 2007 Jun 29;370(1):128-41. doi: 10.1016/j.jmb.2007.04.050. Epub 2007 May 4. J Mol Biol. 2007. PMID: 17512006
-
Crystal structure of a dimeric archaeal splicing endonuclease.J Mol Biol. 2000 Sep 22;302(3):639-48. doi: 10.1006/jmbi.2000.3941. J Mol Biol. 2000. PMID: 10986124
-
RNA-splicing endonuclease structure and function.Cell Mol Life Sci. 2008 Apr;65(7-8):1176-85. doi: 10.1007/s00018-008-7393-y. Cell Mol Life Sci. 2008. PMID: 18217203 Free PMC article. Review.
-
Archaeal introns: splicing, intercellular mobility and evolution.Trends Biochem Sci. 1997 Sep;22(9):326-31. doi: 10.1016/s0968-0004(97)01113-4. Trends Biochem Sci. 1997. PMID: 9301331 Review.
Cited by
-
The life and times of a tRNA.RNA. 2023 Jul;29(7):898-957. doi: 10.1261/rna.079620.123. Epub 2023 Apr 13. RNA. 2023. PMID: 37055150 Free PMC article. Review.
-
Functional importance of crenarchaea-specific extra-loop revealed by an X-ray structure of a heterotetrameric crenarchaeal splicing endonuclease.Nucleic Acids Res. 2009 Aug;37(14):4787-98. doi: 10.1093/nar/gkp506. Epub 2009 Jun 10. Nucleic Acids Res. 2009. PMID: 19515941 Free PMC article.
-
Circularly permuted tRNA genes: their expression and implications for their physiological relevance and development.Front Genet. 2014 Apr 1;5:63. doi: 10.3389/fgene.2014.00063. eCollection 2014. Front Genet. 2014. PMID: 24744771 Free PMC article. Review.
-
X-ray structure of the fourth type of archaeal tRNA splicing endonuclease: insights into the evolution of a novel three-unit composition and a unique loop involved in broad substrate specificity.Nucleic Acids Res. 2012 Nov 1;40(20):10554-66. doi: 10.1093/nar/gks826. Epub 2012 Aug 31. Nucleic Acids Res. 2012. PMID: 22941657 Free PMC article.
-
A novel three-unit tRNA splicing endonuclease found in ultrasmall Archaea possesses broad substrate specificity.Nucleic Acids Res. 2011 Dec;39(22):9695-704. doi: 10.1093/nar/gkr692. Epub 2011 Aug 31. Nucleic Acids Res. 2011. PMID: 21880595 Free PMC article.
References
-
- Abelson J, Trotta CR, Li H. J Biol Chem. 1998;273:12685–12688. - PubMed
-
- Belfort M, Weiner A. Cell. 1997;89:1003–1006. - PubMed
-
- Reyes VM, Abelson J. Cell. 1988;55:719–730. - PubMed
-
- Mattoccia E, Baldi IM, Gandini-Attardi D, Ciafre S, Tocchini-Valentini GP. Cell. 1988;55:731–738. - PubMed
-
- Di Nicola Negri E, Fabbri S, Bufardeci E, Baldi MI, Gandini Attardi D, Mattoccia E, Tocchini-Valentini GP. Cell. 1997;89:859–866. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

