Conformational entropy in molecular recognition by proteins
- PMID: 17637663
- PMCID: PMC4156320
- DOI: 10.1038/nature05959
Conformational entropy in molecular recognition by proteins
Abstract
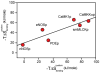
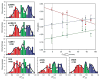
Molecular recognition by proteins is fundamental to almost every biological process, particularly the protein associations underlying cellular signal transduction. Understanding the basis for protein-protein interactions requires the full characterization of the thermodynamics of their association. Historically it has been virtually impossible to experimentally estimate changes in protein conformational entropy, a potentially important component of the free energy of protein association. However, nuclear magnetic resonance spectroscopy has emerged as a powerful tool for characterizing the dynamics of proteins. Here we employ changes in conformational dynamics as a proxy for corresponding changes in conformational entropy. We find that the change in internal dynamics of the protein calmodulin varies significantly on binding a variety of target domains. Surprisingly, the apparent change in the corresponding conformational entropy is linearly related to the change in the overall binding entropy. This indicates that changes in protein conformational entropy can contribute significantly to the free energy of protein-ligand association.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures

References
-
- Wodak SJ, Janin J. Structural basis of macromolecular recognition. Adv Prot Chem. 2002;61:9–73. - PubMed
-
- Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. - PubMed
-
- Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

