Modeling insertional mutagenesis using gene length and expression in murine embryonic stem cells
- PMID: 17637833
- PMCID: PMC1910612
- DOI: 10.1371/journal.pone.0000617
Modeling insertional mutagenesis using gene length and expression in murine embryonic stem cells
Abstract
Background: High-throughput mutagenesis of the mammalian genome is a powerful means to facilitate analysis of gene function. Gene trapping in embryonic stem cells (ESCs) is the most widely used form of insertional mutagenesis in mammals. However, the rules governing its efficiency are not fully understood, and the effects of vector design on the likelihood of gene-trapping events have not been tested on a genome-wide scale.
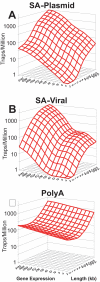
Methodology/principal findings: In this study, we used public gene-trap data to model gene-trap likelihood. Using the association of gene length and gene expression with gene-trap likelihood, we constructed spline-based regression models that characterize which genes are susceptible and which genes are resistant to gene-trapping techniques. We report results for three classes of gene-trap vectors, showing that both length and expression are significant determinants of trap likelihood for all vectors. Using our models, we also quantitatively identified hotspots of gene-trap activity, which represent loci where the high likelihood of vector insertion is controlled by factors other than length and expression. These formalized statistical models describe a high proportion of the variance in the likelihood of a gene being trapped by expression-dependent vectors and a lower, but still significant, proportion of the variance for vectors that are predicted to be independent of endogenous gene expression.
Conclusions/significance: The findings of significant expression and length effects reported here further the understanding of the determinants of vector insertion. Results from this analysis can be applied to help identify other important determinants of this important biological phenomenon and could assist planning of large-scale mutagenesis efforts.
Conflict of interest statement
Figures
Similar articles
-
Enhanced gene trapping in mouse embryonic stem cells.Nucleic Acids Res. 2008 Nov;36(20):e133. doi: 10.1093/nar/gkn603. Epub 2008 Sep 23. Nucleic Acids Res. 2008. PMID: 18812397 Free PMC article.
-
Gene-trap vectors and mutagenesis.Methods Mol Biol. 2009;530:29-47. doi: 10.1007/978-1-59745-471-1_3. Methods Mol Biol. 2009. PMID: 19266330
-
Gene trap: knockout on the fast lane.Methods Mol Biol. 2009;561:145-59. doi: 10.1007/978-1-60327-019-9_10. Methods Mol Biol. 2009. PMID: 19504070
-
International Gene Trap Project: towards gene-driven saturation mutagenesis in mice.Curr Pharm Biotechnol. 2009 Feb;10(2):221-9. doi: 10.2174/138920109787315006. Curr Pharm Biotechnol. 2009. PMID: 19199955 Review.
-
Mouse gene trap approach: identification of novel genes and characterization of their biological functions.Biochem Cell Biol. 1998;76(6):1029-37. Biochem Cell Biol. 1998. PMID: 10392714 Review.
Cited by
-
In vivo protein trapping produces a functional expression codex of the vertebrate proteome.Nat Methods. 2011 Jun;8(6):506-15. doi: 10.1038/nmeth.1606. Epub 2011 May 8. Nat Methods. 2011. PMID: 21552255 Free PMC article.
-
Beyond knockouts: the International Knockout Mouse Consortium delivers modular and evolving tools for investigating mammalian genes.Mamm Genome. 2015 Oct;26(9-10):456-66. doi: 10.1007/s00335-015-9598-3. Epub 2015 Sep 4. Mamm Genome. 2015. PMID: 26340938
-
Efficient conditional and promoter-specific in vivo expression of cDNAs of choice by taking advantage of recombinase-mediated cassette exchange using FlEx gene traps.Nucleic Acids Res. 2010 May;38(9):e106. doi: 10.1093/nar/gkq044. Epub 2010 Feb 5. Nucleic Acids Res. 2010. PMID: 20139417 Free PMC article.
-
Genome Wide Conditional Mouse Knockout Resources.Drug Discov Today Dis Models. 2016 Summer;20:3-12. doi: 10.1016/j.ddmod.2017.08.002. Epub 2017 Sep 12. Drug Discov Today Dis Models. 2016. PMID: 39132094 Free PMC article.
-
Enhanced gene trapping in mouse embryonic stem cells.Nucleic Acids Res. 2008 Nov;36(20):e133. doi: 10.1093/nar/gkn603. Epub 2008 Sep 23. Nucleic Acids Res. 2008. PMID: 18812397 Free PMC article.
References
-
- Matthews K, Kaufman T, Gelbart W. Research resources for Drosophila: the expanding universe. Nat Rev Genet. 2005;6(3):179–193. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

