Primate TNF promoters reveal markers of phylogeny and evolution of innate immunity
- PMID: 17637837
- PMCID: PMC1905939
- DOI: 10.1371/journal.pone.0000621
Primate TNF promoters reveal markers of phylogeny and evolution of innate immunity
Abstract
Background: Tumor necrosis factor (TNF) is a critical cytokine in the immune response whose transcriptional activation is controlled by a proximal promoter region that is highly conserved in mammals and, in particular, primates. Specific single nucleotide polymorphisms (SNPs) upstream of the proximal human TNF promoter have been identified, which are markers of human ancestry.
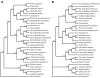
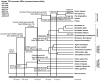
Methodology/principal findings: Using a comparative genomics approach we show that certain fixed genetic differences in the TNF promoter serve as markers of primate speciation. We also demonstrate that distinct alleles of most human TNF promoter SNPs are identical to fixed nucleotides in primate TNF promoters. Furthermore, we identify fixed genetic differences within the proximal TNF promoters of Asian apes that do not occur in African ape or human TNF promoters. Strikingly, protein-DNA binding assays and gene reporter assays comparing these Asian ape TNF promoters to African ape and human TNF promoters demonstrate that, unlike the fixed differences that we define that are associated with primate phylogeny, these Asian ape-specific fixed differences impair transcription factor binding at an Sp1 site and decrease TNF transcription induced by bacterial stimulation of macrophages.
Conclusions/significance: Here, we have presented the broadest interspecies comparison of a regulatory region of an innate immune response gene to date. We have characterized nucleotide positions in Asian ape TNF promoters that underlie functional changes in cell type- and stimulus-specific activation of the TNF gene. We have also identified ancestral TNF promoter nucleotide states in the primate lineage that correspond to human SNP alleles. These findings may reflect evolution of Asian and African apes under a distinct set of infectious disease pressures involving the innate immune response and TNF.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

