Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome
- PMID: 17638852
- PMCID: PMC2018675
- DOI: 10.1182/blood-2006-12-061507
Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome
Erratum in
- Blood. 2008 Jan 1;111(1):49
Abstract
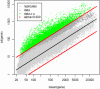
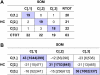
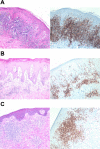
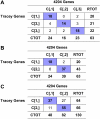
Cutaneous T-cell lymphoma (CTCL) is defined by infiltration of activated and malignant T cells in the skin. The clinical manifestations and prognosis in CTCL are highly variable. In this study, we hypothesized that gene expression analysis in lesional skin biopsies can improve understanding of the disease and its management. Based on 63 skin samples, we performed consensus clustering, revealing 3 patient clusters. Of these, 2 clusters tended to differentiate limited CTCL (stages IA and IB) from more extensive CTCL (stages IB and III). Stage IB patients appeared in both clusters, but those in the limited CTCL cluster were more responsive to treatment than those in the more extensive CTCL cluster. The third cluster was enriched in lymphocyte activation genes and was associated with a high proportion of tumor (stage IIB) lesions. Survival analysis revealed significant differences in event-free survival between clusters, with poorest survival seen in the activated lymphocyte cluster. Using supervised analysis, we further characterized genes significantly associated with lower-stage/treatment-responsive CTCL versus higher-stage/treatment-resistant CTCL. We conclude that transcriptional profiling of CTCL skin lesions reveals clinically relevant signatures, correlating with differences in survival and response to treatment. Additional prospective long-term studies to validate and refine these findings appear warranted.
Figures





Similar articles
-
Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma.Clin Cancer Res. 2010 Apr 1;16(7):2106-14. doi: 10.1158/1078-0432.CCR-09-2879. Epub 2010 Mar 16. Clin Cancer Res. 2010. PMID: 20233883 Free PMC article.
-
Single-Cell Lymphocyte Heterogeneity in Advanced Cutaneous T-cell Lymphoma Skin Tumors.Clin Cancer Res. 2019 Jul 15;25(14):4443-4454. doi: 10.1158/1078-0432.CCR-19-0148. Epub 2019 Apr 22. Clin Cancer Res. 2019. PMID: 31010835 Free PMC article.
-
Primary T Cells from Cutaneous T-cell Lymphoma Skin Explants Display an Exhausted Immune Checkpoint Profile.Cancer Immunol Res. 2018 Aug;6(8):900-909. doi: 10.1158/2326-6066.CIR-17-0270. Epub 2018 Jun 12. Cancer Immunol Res. 2018. PMID: 29895574 Free PMC article.
-
Comparative study of cutaneous T-cell lymphoma and adult T-cell leukemia/lymphoma. Clinical, histopathologic, and immunohistochemical analyses.Cancer. 1990 Dec 1;66(11):2380-6. doi: 10.1002/1097-0142(19901201)66:11<2380::aid-cncr2820661122>3.0.co;2-h. Cancer. 1990. PMID: 2245393 Review.
-
Insights Into the Molecular and Cellular Underpinnings of Cutaneous T Cell Lymphoma.Yale J Biol Med. 2020 Mar 27;93(1):111-121. eCollection 2020 Mar. Yale J Biol Med. 2020. PMID: 32226341 Free PMC article. Review.
Cited by
-
The expression of Eps15 homology domain 1 is negatively correlated with disease-free survival and overall survival of osteosarcoma patients.J Orthop Surg Res. 2019 Apr 11;14(1):103. doi: 10.1186/s13018-019-1137-6. J Orthop Surg Res. 2019. PMID: 30975166 Free PMC article.
-
Pilot Study of a Novel Therapeutic Approach for Refractory Advanced Stage Folliculotropic Mycosis Fungoides.Acta Derm Venereol. 2020 Jun 18;100(13):adv00187. doi: 10.2340/00015555-3443. Acta Derm Venereol. 2020. PMID: 32128597 Free PMC article.
-
Increased Expression of Eps15 Homology Domain 1 is Associated with Poor Prognosis in Resected Small Cell Lung Cancer.J Cancer. 2015 Aug 20;6(10):990-5. doi: 10.7150/jca.11650. eCollection 2015. J Cancer. 2015. PMID: 26366212 Free PMC article.
-
Re-activation of mitochondrial apoptosis inhibits T-cell lymphoma survival and treatment resistance.Leukemia. 2016 Jul;30(7):1520-30. doi: 10.1038/leu.2016.49. Epub 2016 Mar 8. Leukemia. 2016. PMID: 27055871
-
Role of B-cells in Mycosis Fungoides.Acta Derm Venereol. 2021 Mar 11;101(3):adv00413. doi: 10.2340/00015555-3775. Acta Derm Venereol. 2021. PMID: 33686443 Free PMC article.
References
-
- Shin J, Jones DA, Hurwitz D, Kupper TS. Gene expression profiling in cutaneous T cell lymphoma skin lesions. J Invest Dermatol. 2006;126(Supplement):24.
-
- Willemze R, Jaffe ES, Burg G, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005;105:3768–3785. - PubMed
-
- Zackheim HS, Amin S, Kashani-Sabet M, McMillan A. Prognosis in cutaneous T-cell lymphoma by skin stage: long-term survival in 489 patients. J Am Acad Dermatol. 1999;40:418–425. - PubMed
-
- Diamandidou E, Colome-Grimmer M, Fayad L, Duvic M, Kurzrock R. Transformation of mycosis fungoides/Sezary syndrome: clinical characteristics and prognosis. Blood. 1998;92:1150–1159. - PubMed
-
- Kim YH, Liu HL, Mraz-Gernhard S, Varghese A, Hoppe RT. Long-term outcome of 525 patients with mycosis fungoides and Sezary syndrome: clinical prognostic factors and risk for disease progression. Arch Dermatol. 2003;139:857–866. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

