The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3
- PMID: 17638854
- PMCID: PMC2200917
- DOI: 10.1182/blood-2007-06-094748
The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3
Abstract
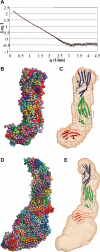
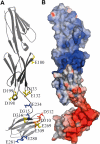
The Lutheran blood group glycoprotein, first discovered on erythrocytes, is widely expressed in human tissues. It is a ligand for the alpha5 subunit of Laminin 511/521, an extracellular matrix protein. This interaction may contribute to vaso-occlusive events that are an important cause of morbidity in sickle cell disease. Using x-ray crystallography, small-angle x-ray scattering, and site-directed mutagenesis, we show that the extracellular region of Lutheran forms an extended structure with a distinctive bend between the second and third immunoglobulin-like domains. The linker between domains 2 and 3 appears to be flexible and is a critical determinant in maintaining an overall conformation for Lutheran that is capable of binding to Laminin. Mutagenesis studies indicate that Asp312 of Lutheran and the surrounding cluster of negatively charged residues in this linker region form the Laminin-binding site. Unusually, receptor binding is therefore not a function of the domains expected to be furthermost from the plasma membrane. These studies imply that structural flexibility of Lutheran may be essential for its interaction with Laminin and present a novel opportunity for the development of therapeutics for sickle cell disease.
Figures
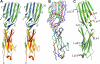


References
-
- Parsons SF, Mallinson G, Judson PA, et al. Evidence that the Lub blood group antigen is located on red cell membrane glycoproteins of 85 and 78 kd. Transfusion. 1987;27:61–63. - PubMed
-
- Campbell IG, Foulkes WD, Senger G, et al. Molecular cloning of the B-CAM cell surface glycoprotein of epithelial cancers: a novel member of the immunoglobulin superfamily. Cancer Res. 1994;54:5761–5765. - PubMed
-
- Rahuel C, Le Van KC, Mattei MG, Cartron JP, Colin Y. A unique gene encodes spliceoforms of the B-cell adhesion molecule cell surface glycoprotein of epithelial cancer and of the Lutheran blood group glycoprotein. Blood. 1996;88:1865–1872. - PubMed
-
- El Nemer W, Colin Y, Bauvy C, et al. Isoforms of the Lutheran/basal cell adhesion molecule glycoprotein are differentially delivered in polarized epithelial cells: mapping of the basolateral sorting signal to a cytoplasmic di-leucine motif. J Biol Chem. 1999;274:31903–31908. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

