The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli
- PMID: 17640276
- PMCID: PMC2170320
- DOI: 10.1111/j.1365-2958.2007.05851.x
The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli
Abstract
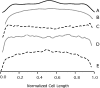
In Escherichia coli many enzymes including MurG are directly involved in the synthesis and assembly of peptidoglycan. MurG is an essential glycosyltransferase catalysing the last intracellular step of peptidoglycan synthesis. To elucidate its role during elongation and division events, localization of MurG using immunofluorescence microscopy was performed. MurG exhibited a random distribution in the cell envelope with a relatively higher intensity at the division site. This mid-cell localization was dependent on the presence of a mature divisome. Its localization in the lateral cell wall appeared to require the presence of MreCD. This could be indicative of a potential interaction between MurG and other proteins. Investigating this by immunoprecipitation revealed the association of MurG with MreB and MraY in the same protein complex. In view of this, the loss of rod shape of DeltamreBCD strain could be ascribed to the loss of MurG membrane localization. Consequently, this could prevent the localized supply of the lipid II precursor to the peptidoglycan synthesizing machinery involved in cell elongation. It is postulated that the involvement of MurG in the peptidoglycan synthesis concurs with two complexes, one implicated in cell elongation and the other in division. A model representing the first complex is proposed.
Figures







Similar articles
-
Structural determinants required to target penicillin-binding protein 3 to the septum of Escherichia coli.J Bacteriol. 2004 Sep;186(18):6110-7. doi: 10.1128/JB.186.18.6110-6117.2004. J Bacteriol. 2004. PMID: 15342580 Free PMC article.
-
Assay for identification of inhibitors for bacterial MraY translocase or MurG transferase.Anal Biochem. 2000 May 1;280(2):315-9. doi: 10.1006/abio.2000.4530. Anal Biochem. 2000. PMID: 10790316
-
The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus.Mol Microbiol. 2007 May;64(4):938-52. doi: 10.1111/j.1365-2958.2007.05720.x. Mol Microbiol. 2007. PMID: 17501919
-
Morphogenesis of rod-shaped sacculi.FEMS Microbiol Rev. 2008 Mar;32(2):321-44. doi: 10.1111/j.1574-6976.2007.00090.x. FEMS Microbiol Rev. 2008. PMID: 18291013 Review.
-
The biosynthesis of peptidoglycan lipid-linked intermediates.FEMS Microbiol Rev. 2008 Mar;32(2):208-33. doi: 10.1111/j.1574-6976.2007.00089.x. Epub 2007 Dec 10. FEMS Microbiol Rev. 2008. PMID: 18081839 Review.
Cited by
-
Putative hexameric glycosyltransferase functional unit revealed by the crystal structure of Acinetobacter baumannii MurG.IUCrJ. 2021 May 8;8(Pt 4):574-583. doi: 10.1107/S2052252521003729. eCollection 2021 Jul 1. IUCrJ. 2021. PMID: 34258006 Free PMC article.
-
The actin homologue MreB organizes the bacterial cell membrane.Nat Commun. 2014 Mar 7;5:3442. doi: 10.1038/ncomms4442. Nat Commun. 2014. PMID: 24603761 Free PMC article.
-
Flotillin-mediated membrane fluidity controls peptidoglycan synthesis and MreB movement.Elife. 2020 Jul 14;9:e57179. doi: 10.7554/eLife.57179. Elife. 2020. PMID: 32662773 Free PMC article.
-
Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo.PLoS One. 2010 Nov 11;5(11):e13938. doi: 10.1371/journal.pone.0013938. PLoS One. 2010. PMID: 21085642 Free PMC article.
-
Changes of lipid domains in Bacillus subtilis cells with disrupted cell wall peptidoglycan.FEMS Microbiol Lett. 2011 Dec;325(1):92-8. doi: 10.1111/j.1574-6968.2011.02417.x. Epub 2011 Oct 3. FEMS Microbiol Lett. 2011. PMID: 22092867 Free PMC article.
References
-
- Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol Microbiol. 2007;64:938–952. - PubMed
-
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Distèche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. - PubMed
-
- Addinall SG, Cao C, Lutkenhaus J. Temperature shift experiments with an FtsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z Ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–4284. - PMC - PubMed
-
- Auger G, van Heijenoort J, Mengin-Lecreulx D, Blanot D. A MurG assay which utilises a synthetic analogue of lipid I. FEMS Microbiol Lett. 2003;219:115–119. - PubMed
-
- Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, et al. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol Microbiol. 2006;61:675–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

