Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa
- PMID: 17640302
- PMCID: PMC11158818
- DOI: 10.1111/j.1349-7006.2007.00548.x
Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa
Abstract
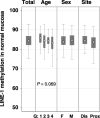
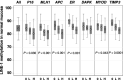
Hypermethylation of CpG island loci within gene promoter regions is a frequent event in colorectal cancer that is often associated with transcriptional silencing and has been referred to as CIMP+. DNA hypomethylation can occur in concert with CIMP+, although these two phenomena appear not to be related in colorectal cancer. The authors investigated here whether the methylation level of LINE-1 repeats, a surrogate marker for genomic methylation, was associated with the level of CpG island methylation in colorectal cancers and in matching normal colonic mucosa from 178 patients. The MethyLight assay was used to quantitate the methylation of CpG islands within the MLH1, P16(INK4A), TIMP3, DAPK, APC, ER and MYOD genes. A real-time, methylation-specific polymerase chain reaction assay was also used to quantitate the methylation of LINE-1 repeats. In colorectal cancer, no associations were seen between methylation levels in LINE-1 repeats and CpG island loci, including a new CpG island panel that was recently proposed for CIMP+. In normal colonic mucosa, however, the methylation level of LINE-1 repeats was inversely correlated with CpG-island methylation of the MLH1, P16, TIMP3, APC, ER and MYOD genes. The methylation level of LINE-1 repeats in normal colonic mucosa also showed significant associations with common polymorphisms in the methylene tetrahydrofolate reductase and methylene tetrahydrofolate dehydrogenase genes involved in methyl group metabolism. Further investigation of genomic and CpG island methylation in normal colonic mucosa and the possible influences of environmental and genetic factors may provide new insights into the development of CIMP+ colorectal cancer.
Figures


Similar articles
-
DNA hypermethylation in the normal colonic mucosa of patients with colorectal cancer.Br J Cancer. 2006 Feb 27;94(4):593-8. doi: 10.1038/sj.bjc.6602940. Br J Cancer. 2006. PMID: 16421593 Free PMC article.
-
DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer.Clin Cancer Res. 2009 Jun 1;15(11):3663-71. doi: 10.1158/1078-0432.CCR-08-2383. Epub 2009 May 26. Clin Cancer Res. 2009. PMID: 19470733 Free PMC article.
-
LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer.Int J Cancer. 2008 Jun 15;122(12):2767-73. doi: 10.1002/ijc.23470. Int J Cancer. 2008. PMID: 18366060 Free PMC article.
-
APC gene methylation is inversely correlated with features of the CpG island methylator phenotype in colorectal cancer.Int J Cancer. 2006 Nov 15;119(10):2272-8. doi: 10.1002/ijc.22237. Int J Cancer. 2006. PMID: 16981189
-
Epigenetic alterations in colorectal cancer: the CpG island methylator phenotype.Histol Histopathol. 2013 May;28(5):585-95. doi: 10.14670/HH-28.585. Epub 2013 Jan 23. Histol Histopathol. 2013. PMID: 23341177 Review.
Cited by
-
Association of Methylenetetrahydrofolate Reductase rs1801133 Gene Polymorphism with Cancer Risk and Septin 9 Methylation in Patients with Colorectal Cancer.J Gastrointest Cancer. 2024 Jun;55(2):778-786. doi: 10.1007/s12029-024-01020-y. Epub 2024 Jan 22. J Gastrointest Cancer. 2024. PMID: 38252186 Free PMC article.
-
Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease.Mod Pathol. 2013 Apr;26(4):465-84. doi: 10.1038/modpathol.2012.214. Epub 2013 Jan 11. Mod Pathol. 2013. PMID: 23307060 Free PMC article. Review.
-
Long interspersed nuclear element-1 hypomethylation in cancer: biology and clinical applications.Clin Epigenetics. 2011 Aug;2(2):315-30. doi: 10.1007/s13148-011-0032-8. Epub 2011 Apr 10. Clin Epigenetics. 2011. PMID: 22704344 Free PMC article.
-
LINE-1 Hypermethylation in Serum Cell-Free DNA of Relapsing Remitting Multiple Sclerosis Patients.Mol Neurobiol. 2018 Jun;55(6):4681-4688. doi: 10.1007/s12035-017-0679-z. Epub 2017 Jul 13. Mol Neurobiol. 2018. PMID: 28707075 Free PMC article.
-
Concordance of Hypermethylated DNA and the Tumor Markers CA 15-3, CEA, and TPA in Serum during Monitoring of Patients with Advanced Breast Cancer.Biomed Res Int. 2015;2015:986024. doi: 10.1155/2015/986024. Epub 2015 Aug 3. Biomed Res Int. 2015. PMID: 26339655 Free PMC article.
References
-
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3: 415–28. - PubMed
-
- Laird PW. Cancer epigenetics. Hum Mol Genet 2005; 14: R65–76. - PubMed
-
- Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene 2002; 21: 5400–13. - PubMed
-
- Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumours promoted by DNA hypomethylation. Science 2003; 300: 455. - PubMed
-
- Matsuzaki K, Deng G, Tanaka H et al . The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res 2005; 11: 8564–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

