The correlation between architecture and mRNA abundance in the genetic regulatory network of Escherichia coli
- PMID: 17640329
- PMCID: PMC1940267
- DOI: 10.1186/1752-0509-1-30
The correlation between architecture and mRNA abundance in the genetic regulatory network of Escherichia coli
Abstract
Background: Two aspects of genetic regulatory networks are the static architecture that describes the overall connectivity between the genes and the dynamics that describes the sequence of genes active at any one time as deduced from mRNA abundances. The nature of the relationship between these two aspects of these networks is a fundamental question. To address it, we have used the static architecture of the connectivity of the regulatory proteins of Escherichia coli to analyse their relationship to the abundance of the mRNAs encoding these proteins. In this we build on previous work which uses Boolean network models, but impose biological constraints that cannot be deduced from the mRNA abundances alone.
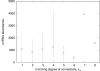
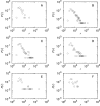
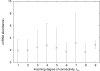
Results: For a cell population of E. coli, we find that there is a strong and statistically significant linear dependence between the abundance of mRNA encoding a regulatory protein and the number of genes regulated by this protein. We use this result, together with the ratio of regulatory repressors to promoters, to simulate numerically a genetic regulatory network of a single cell. The resulting model exhibits similar correlations to that of E. coli.
Conclusion: This analysis clarifies the relationship between the static architecture of a regulatory network and the consequences for the dynamics of its pattern of mRNA abundances. It also provides the constraints on the architecture required to construct a model network to simulate mRNA production.
Figures


References
-
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. - DOI - PubMed
-
- Raine DJ, Norris V. Network Structure of Metabolic Pathways. InterJournal of Complex Systems. 2000.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

