MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress
- PMID: 17640358
- PMCID: PMC1947970
- DOI: 10.1186/1471-2164-8-242
MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress
Abstract
Background: MADS-box transcription factors, besides being involved in floral organ specification, have also been implicated in several aspects of plant growth and development. In recent years, there have been reports on genomic localization, protein motif structure, phylogenetic relationships, gene structure and expression of the entire MADS-box family in the model plant system, Arabidopsis. Though there have been some studies in rice as well, an analysis of the complete MADS-box family along with a comprehensive expression profiling was still awaited after the completion of rice genome sequencing. Furthermore, owing to the role of MADS-box family in flower development, an analysis involving structure, expression and functional aspects of MADS-box genes in rice and Arabidopsis was required to understand the role of this gene family in reproductive development.
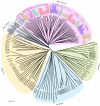
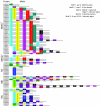
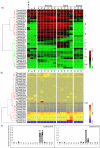
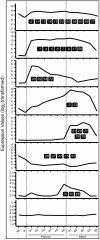
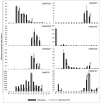
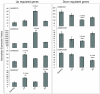
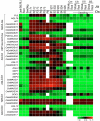
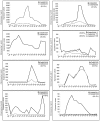
Results: A genome-wide molecular characterization and microarray-based expression profiling of the genes encoding MADS-box transcription factor family in rice is presented. Using a thorough annotation exercise, 75 MADS-box genes have been identified in rice and categorized into MIKCc, MIKC*, Malpha, Mbeta and Mgamma groups based on phylogeny. Chromosomal localization of these genes reveals that 16 MADS-box genes, mostly MIKCc-type, are located within the duplicated segments of the rice genome, whereas most of the M-type genes, 20 in all, seem to have resulted from tandem duplications. Nine members belonging to the Mbeta group, which was considered absent in monocots, have also been identified. The expression profiles of all the MADS-box genes have been analyzed under 11 temporal stages of panicle and seed development, three abiotic stress conditions, along with three stages of vegetative development. Transcripts for 31 genes accumulate preferentially in the reproductive phase, of which, 12 genes are specifically expressed in seeds, and six genes show expression specific to panicle development. Differential expression of seven genes under stress conditions is also evident. An attempt has been made to gain insight into plausible functions of rice MADS-box genes by collating the expression data of functionally validated genes in rice and Arabidopsis.
Conclusion: Only a limited number of MADS genes have been functionally validated in rice. A comprehensive annotation and transcriptome profiling undertaken in this investigation adds to our understanding of the involvement of MADS-box family genes during reproductive development and stress in rice and also provides the basis for selection of candidate genes for functional validation studies.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

