The function and regulation of Ultrabithorax in the legs of Drosophila melanogaster
- PMID: 17640629
- PMCID: PMC2040266
- DOI: 10.1016/j.ydbio.2007.06.002
The function and regulation of Ultrabithorax in the legs of Drosophila melanogaster
Abstract
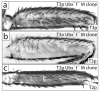
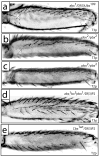
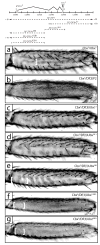
Alterations in Hox gene expression patterns have been implicated in both large and small-scale morphological evolution. An improved understanding of these changes requires a detailed understanding of Hox gene cis-regulatory function and evolution. cis-regulatory evolution of the Hox gene Ultrabithorax (Ubx) has been shown to contribute to evolution of trichome patterns on the posterior second femur (T2p) of Drosophila species. As a step toward determining how this function of Ubx has evolved, we performed a series of experiments to clarify the role of Ubx in patterning femurs and to identify the cis-regulatory regions of Ubx that drive expression in T2p. We first performed clonal analysis to further define Ubx function in patterning bristle and trichome patterns in the legs. We found that low levels of Ubx expression are sufficient to repress an eighth bristle row on the posterior second and third femurs, whereas higher levels of expression are required to promote the development and migration of other bristles on the third femur and to repress trichomes. We then tested the hypothesis that the evolutionary difference in T2p trichome patterns due to Ubx was caused by a change in the global cis-regulation of Ubx expression. We found no evidence to support this view, suggesting that the evolved difference in Ubx function reflects evolution of a leg-specific enhancer. We then searched for the regulatory regions of the Ubx locus that drive expression in the second and third femur by assaying all existing regulatory mutations of the Ubx locus and new deficiencies in the large intron of Ubx that we generated by P-element-induced male recombination. We found that two enhancer regions previously known to regulate Ubx expression in the legs, abx and pbx, are required for Ubx expression in the third femur, but that they do not contribute to pupal expression of Ubx in the second femur. This analysis allowed us to rule out at least 100 kb of DNA in and around the Ubx locus as containing a T2p-specific enhancer. We then surveyed an additional approximately 30 kb using enhancer constructs. None of these enhancer constructs produced an expression pattern similar to Ubx expression in T2p. Thus, after surveying over 95% of the Ubx locus, we have not been able to localize a T2p-specific enhancer. While the enhancer could reside within the small regions we have not surveyed, it is also possible that the enhancer is structurally complex and/or acts only within its native genomic context.
Figures




References
-
- Ashburner M. Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989.
-
- Averof M, Patel NH. Crustacean appendage evolution associated with changes in Hox gene expression. Nature. 1997;388:682–686. - PubMed
-
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726, 728. 2000;730:732. - PubMed
-
- Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. - PubMed
-
- Bienz M, Saari G, Tremml G, Müller J, Züst B, Lawrence PA. Differential regulation of Ultrabithorax in two germ layers of Drosophila. Cell. 1988;53:567–576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

