Structural analysis and dynamics of retinal chromophore in dark and meta I states of rhodopsin from 2H NMR of aligned membranes
- PMID: 17640664
- PMCID: PMC5233725
- DOI: 10.1016/j.jmb.2007.03.046
Structural analysis and dynamics of retinal chromophore in dark and meta I states of rhodopsin from 2H NMR of aligned membranes
Abstract
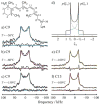
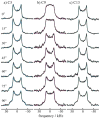
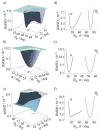
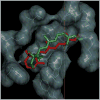
Rhodopsin is a prototype for G protein-coupled receptors (GPCRs) that are implicated in many biological responses in humans. A site-directed (2)H NMR approach was used for structural analysis of retinal within its binding cavity in the dark and pre-activated meta I states. Retinal was labeled with (2)H at the C5, C9, or C13 methyl groups by total synthesis, and was used to regenerate the opsin apoprotein. Solid-state (2)H NMR spectra were acquired for aligned membranes in the low-temperature lipid gel phase versus the tilt angle to the magnetic field. Data reduction assumed a static uniaxial distribution, and gave the retinylidene methyl bond orientations plus the alignment disorder (mosaic spread). The dark-state (2)H NMR structure of 11-cis-retinal shows torsional twisting of the polyene chain and the beta-ionone ring. The ligand undergoes restricted motion, as evinced by order parameters of approximately 0.9 for the spinning C-C(2)H(3) groups, with off-axial fluctuations of approximately 15 degrees . Retinal is accommodated within the rhodopsin binding pocket with a negative pre-twist about the C11=C12 double bond that explains its rapid photochemistry and the trajectory of 11-cis to trans isomerization. In the cryo-trapped meta I state, the (2)H NMR structure shows a reduction of the polyene strain, while torsional twisting of the beta-ionone ring is maintained. Distortion of the retinal conformation is interpreted through substituent control of receptor activation. Steric hindrance between trans retinal and Trp265 can trigger formation of the subsequent activated meta II state. Our results are pertinent to quantum and molecular mechanics simulations of ligands bound to GPCRs, and illustrate how (2)H NMR can be applied to study their biological mechanisms of action.
Figures









References
-
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. - PubMed
-
- Fanelli F, De Benedetti PG. Computational modeling approaches to structure-function analysis of G protein-coupled receptors. Chem Rev. 2005;105:3297–3351. - PubMed
-
- Sakmar TP, Menon ST, Marin EP, Awad ES. Rhodopsin: Insights from recent structural studies. Annu Rev Biophys Biomol Struct. 2002;31:443–484. - PubMed
-
- Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Prot Chem. 2003;63:243–290. - PubMed
-
- Lüdeke S, Beck R, Yan ECY, Sakmar TP, Siebert F, Vogel R. The role of Glu181 in the photoactivation of rhodopsin. J Mol Biol. 2005;353:345–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

