Widely distributed noncoding purifying selection in the human genome
- PMID: 17640883
- PMCID: PMC1941483
- DOI: 10.1073/pnas.0705140104
Widely distributed noncoding purifying selection in the human genome
Abstract
It is widely assumed that human noncoding sequences comprise a substantial reservoir for functional variants impacting gene regulation and other chromosomal processes. Evolutionarily conserved noncoding sequences (CNSs) in the human genome have attracted considerable attention for their potential to simplify the search for functional elements and phenotypically important human alleles. A major outstanding question is whether functionally significant human noncoding variation is concentrated in CNSs or distributed more broadly across the genome. Here, we combine whole genome sequence data from four nonhuman species (chimp, dog, mouse, and rat) with recently available comprehensive human polymorphism data to analyze selection at single-nucleotide resolution. We show that a substantial fraction of active purifying selection in human noncoding sequences occurs outside of CNSs and is diffusely distributed across the genome. This finding suggests the existence of a large complement of human noncoding variants that may impact gene expression and phenotypic traits, the majority of which will escape detection with current approaches to genome analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
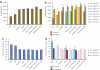

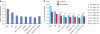
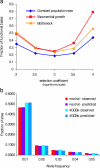
References
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. Science. 2001;291:1304–1351. - PubMed
-
- Miller W, Makova KD, Nekrutenko A, Hardison RC. Annu Rev Genomics Hum Genet. 2004;5:15–56. - PubMed
-
- Boffelli D, Nobrega MA, Rubin EM. Nat Rev Genet. 2004;5:456–465. - PubMed
-
- Dermitzakis ET, Reymond A, Antonarakis SE. Nat Rev Genet. 2005;6:151–157. - PubMed
-
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Nature. 2002;420:520–562. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

