Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion
- PMID: 17640891
- PMCID: PMC1941510
- DOI: 10.1073/pnas.0701737104
Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion
Abstract
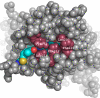
Bacteriophytochromes RpBphP2 and RpBphP3 from the photosynthetic bacterium Rhodopseudomonas palustris work in tandem to modulate synthesis of the light-harvesting complex LH4 in response to light. Although RpBphP2 and RpBphP3 share the same domain structure with 52% sequence identity, they demonstrate distinct photoconversion behaviors. RpBphP2 exhibits the "classical" phytochrome behavior of reversible photoconversion between red (Pr) and far-red (Pfr) light-absorbing states, whereas RpBphP3 exhibits novel photoconversion between Pr and a near-red (Pnr) light-absorbing states. We have determined the crystal structure at 2.2-A resolution of the chromophore binding domains of RpBphP3, covalently bound with chromophore biliverdin IXalpha. By combining structural and sequence analyses with site-directed mutagenesis, we identify key residues that directly modulate the photochemical properties of RpBphP3 and RpBphP2. Remarkably, we identify a region spanning residues 207-212 in RpBphP3, in which a single mutation, L207Y, causes this unusual bacteriophytochrome to revert to the classical phenotype that undergoes reversible photoconversion between the Pr and Pfr states. The reverse mutation, Y193L, in the corresponding region in RpBphP2 significantly diminishes the formation of the Pfr state. We propose that residues 207-212 and the spatially adjacent conserved residues, Asp-216 and Tyr-272, interact with the chromophore and form part of the interface between the chromophore binding domains and the PHY domain that modulates photoconversion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
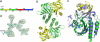


References
-
- Chen E, Lapko VN, Lewis JW, Song PS, Kliger DS. Biochemistry. 1996;35:843–850. - PubMed
-
- Borucki B, von Stetten D, Seibeck S, Lamparter T, Michael N, Mroginski MA, Otto H, Murgida DH, Heyn MP, Hildebrandt P. J Biol Chem. 2005;280:34358–34364. - PubMed
-
- Giraud E, Zappa S, Vuillet L, Adriano JM, Hannibal L, Fardoux J, Berthomieu C, Bouyer P, Pignol D, Vermeglio A. J Biol Chem. 2005;280:32389–32397. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

