ppGpp regulation of RpoS degradation via anti-adaptor protein IraP
- PMID: 17640895
- PMCID: PMC1937563
- DOI: 10.1073/pnas.0705561104
ppGpp regulation of RpoS degradation via anti-adaptor protein IraP
Abstract
IraP is a small protein that interferes with the delivery of sigma(S) (RpoS) to the ClpXP protease by blocking the action of RssB, an adaptor protein for sigma(S) degradation. IraP was previously shown to mediate stabilization of sigma(S) during phosphate starvation. Here, we show that iraP is transcribed in response to phosphate starvation; this response is mediated by ppGpp. The iraP promoter is positively regulated by ppGpp, dependent on the discriminator region of the iraP promoter. Sensing of phosphate starvation requires SpoT but not RelA. The results demonstrate a target for positive regulation by ppGpp and suggest that the cell use of ppGpp to mediate a variety of starvation responses operates in part by modulating sigma(S) levels.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

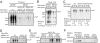
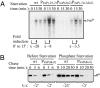
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

