Reevaluating the relationship between EPR spectra and enzyme structure for the iron sulfur clusters in NADH:quinone oxidoreductase
- PMID: 17640900
- PMCID: PMC1925037
- DOI: 10.1073/pnas.0705593104
Reevaluating the relationship between EPR spectra and enzyme structure for the iron sulfur clusters in NADH:quinone oxidoreductase
Abstract
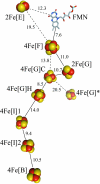
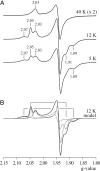
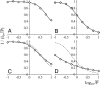
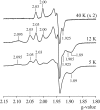
NADH:quinone oxidoreductase (complex I) plays a pivotal role in cellular energy production. It employs a series of redox cofactors to couple electron transfer to the generation of a proton-motive force across the inner mitochondrial or bacterial cytoplasmic membrane. Complex I contains a noncovalently bound flavin mononucleotide at the active site for NADH oxidation and eight or nine iron-sulfur clusters to transfer electrons between the flavin and a quinone-binding site. Understanding the mechanism of complex I requires the properties of these clusters to be defined, both individually and as an ensemble. Most functional information on the clusters has been gained from EPR spectroscopy, but some clusters are not observed by EPR and attributing the observed signals to the structurally defined clusters is difficult. The current consensus picture relies on correlating the spectra from overexpressed subunits (containing one to four clusters) with those from intact complexes I. Here, we analyze spectra from the overexpressed NuoG subunit from Escherichia coli complex I and compare them with spectra from the intact enzyme. Consequently, we propose that EPR signals N4 and N5 have been misassigned: signal N4 is from NuoI (not NuoG) and signal N5 is from the conserved cysteine-ligated [4Fe-4S] cluster in NuoG (not from the cluster with a histidine ligand). The consequences of reassigning the EPR signals and their associated functional information on the free energy profile for electron transfer through complex I are discussed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Walker JE. Q Rev Biophys. 1992;25:253–324. - PubMed
-
- Yagi T, Matsuno-Yagi A. Biochemistry. 2003;42:2266–2274. - PubMed
-
- Hirst J, Carroll J, Fearnley IM, Shannon RJ, Walker JE. Biochim Biophys Acta. 2003;1604:135–150. - PubMed
-
- Sazanov LA. Biochemistry. 2007;46:2275–2288. - PubMed
-
- Hinchliffe P, Sazanov LA. Science. 2005;309:771–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

