High-resolution analysis of Drosophila heterochromatin organization using SuUR Su(var)3-9 double mutants
- PMID: 17640911
- PMCID: PMC1937550
- DOI: 10.1073/pnas.0704690104
High-resolution analysis of Drosophila heterochromatin organization using SuUR Su(var)3-9 double mutants
Abstract
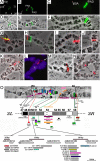
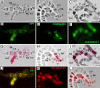
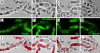
The structural and functional analyses of heterochromatin are essential to understanding how heterochromatic genes are regulated and how centromeric chromatin is formed. Because the repetitive nature of heterochromatin hampers its genome analysis, new approaches need to be developed. Here, we describe how, in double mutants for Su(var)3-9 and SuUR genes encoding two structural proteins of heterochromatin, new banded heterochromatic segments appear in all polytene chromosomes due to the strong suppression of under-replication in pericentric regions. FISH on salivary gland polytene chromosomes from these double mutant larvae allows high resolution of heterochromatin mapping. In addition, immunostaining experiments with a set of antibodies against euchromatic and heterochromatic proteins reveal their unusual combinations in the newly appeared segments: binding patterns for HP1 and HP2 are coincident, but both are distinct from H3diMetK9 and H4triMetK20. In several regions, partial overlapping staining is observed for the proteins characteristic of active chromatin RNA Pol II, H3triMetK4, Z4, and JIL1, the boundary protein BEAF, and the heterochromatin-enriched proteins HP1, HP2, and SU(VAR)3-7. The exact cytological position of the centromere of chromosome 3 was visualized on salivary gland polytene chromosomes by using the centromeric dodeca satellite and the centromeric protein CID. This region is enriched in H3diMetK9 and H4triMetK20 but is devoid of other proteins analyzed.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The SuUR gene influences the distribution of heterochromatic proteins HP1 and SU(VAR)3-9 on nurse cell polytene chromosomes of Drosophila melanogaster.Chromosoma. 2006 Aug;115(4):296-310. doi: 10.1007/s00412-005-0044-2. Epub 2006 Apr 11. Chromosoma. 2006. PMID: 16607511
-
Effects of Mutations in the Drosophila melanogaster Rif1 Gene on the Replication and Underreplication of Pericentromeric Heterochromatin in Salivary Gland Polytene Chromosomes.Cells. 2020 Jun 19;9(6):1501. doi: 10.3390/cells9061501. Cells. 2020. PMID: 32575592 Free PMC article.
-
Microdissection and sequence analysis of pericentric heterochromatin from the Drosophila melanogastermutant Suppressor of Underreplication.Chromosoma. 2002 Jul;111(2):114-25. doi: 10.1007/s00412-002-0190-8. Epub 2002 Apr 16. Chromosoma. 2002. PMID: 12111334
-
Histone modification and the control of heterochromatic gene silencing in Drosophila.Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review.
-
[Contribution of the SuUR gene to the organization of epigenetically repressed regions of Drosophila melanogaster chromosomes].Genetika. 2006 Aug;42(8):1013-28. Genetika. 2006. PMID: 17025152 Review. Russian.
Cited by
-
The SUUR protein is involved in binding of SU(VAR)3-9 and methylation of H3K9 and H3K27 in chromosomes of Drosophila melanogaster.Chromosome Res. 2011 Feb;19(2):235-49. doi: 10.1007/s10577-011-9193-8. Epub 2011 Feb 22. Chromosome Res. 2011. PMID: 21340745
-
Cytological heterogeneity of heterochromatin among 10 sequenced Drosophila species.Genetics. 2022 Sep 30;222(2):iyac119. doi: 10.1093/genetics/iyac119. Genetics. 2022. PMID: 35946576 Free PMC article.
-
Epigenetic Silencing of P-Element Reporter Genes Induced by Transcriptionally Active Domains of Constitutive Heterochromatin in Drosophila melanogaster.Genes (Basel). 2022 Dec 21;14(1):12. doi: 10.3390/genes14010012. Genes (Basel). 2022. PMID: 36672753 Free PMC article.
-
Simple and Complex Centromeric Satellites in Drosophila Sibling Species.Genetics. 2018 Mar;208(3):977-990. doi: 10.1534/genetics.117.300620. Epub 2018 Jan 5. Genetics. 2018. PMID: 29305387 Free PMC article.
-
Intercalary heterochromatin in polytene chromosomes of Drosophila melanogaster.Chromosoma. 2008 Oct;117(5):411-8. doi: 10.1007/s00412-008-0163-7. Epub 2008 May 20. Chromosoma. 2008. PMID: 18491121 Review.
References
-
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, et al. Science. 2000;287:2185–2195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

