Peroxisomal peripheral membrane protein YlInp1p is required for peroxisome inheritance and influences the dimorphic transition in the yeast Yarrowia lipolytica
- PMID: 17644654
- PMCID: PMC2043367
- DOI: 10.1128/EC.00185-07
Peroxisomal peripheral membrane protein YlInp1p is required for peroxisome inheritance and influences the dimorphic transition in the yeast Yarrowia lipolytica
Abstract
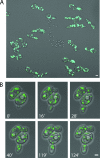
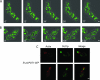
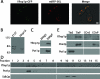
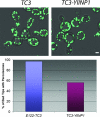
Eukaryotic cells have evolved molecular mechanisms to ensure the faithful inheritance of organelles by daughter cells in order to maintain the benefits afforded by the compartmentalization of biochemical functions. Little is known about the inheritance of peroxisomes, organelles of lipid metabolism. We have analyzed peroxisome dynamics and inheritance in the dimorphic yeast Yarrowia lipolytica. Most peroxisomes are anchored at the periphery of cells of Y. lipolytica. In vivo video microscopy showed that at cell division, approximately half of the anchored peroxisomes in the mother cell are dislodged individually from their static positions and transported to the bud. Peroxisome motility is dependent on the actin cytoskeleton. YlInp1p is a peripheral peroxisomal membrane protein that affects the partitioning of peroxisomes between mother cell and bud in Y. lipolytica. In cells lacking YlInp1p, most peroxisomes were transferred to the bud, with only a few remaining in the mother cell, while in cells overexpressing YlInp1p, peroxisomes were preferentially retained in the mother cell, resulting in buds nearly devoid of peroxisomes. Our results are consistent with a role for YlInp1p in anchoring peroxisomes in cells. YlInp1p has a role in the dimorphic transition in Y. lipolytica, as cells lacking the YlINP1 gene more readily convert from the yeast to the mycelial form in oleic acid-containing medium, the metabolism of which requires peroxisomal activity, than does the wild-type strain. This study reports the first analysis of organelle inheritance in a true dimorphic yeast and identifies the first protein required for peroxisome inheritance in Y. lipolytica.
Figures





References
-
- Davidson, R. C., J. R. Blankership, P. R. Kraus, M. de Jesus Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. - PubMed
-
- de Duve, C., and P. Baudhuin. 1966. Peroxisomes (microbodies and related particles). Physiol. Rev. 46:323-357. - PubMed
-
- Fagarasanu, A., M. Fagarasanu, G. A. Eitzen, J. D. Aitchison, and R. A. Rachubinski. 2006. The peroxisomal membrane protein Inp2p is the peroxisome-specific receptor for the myosin V motor Myo2p of Saccharomyces cerevisiae. Dev. Cell 10:587-600. - PubMed
-
- Fagarasanu, M., A. Fagarasanu, and R. A. Rachubinski. 2006. Sharing the wealth: peroxisome inheritance in budding yeast. Biochim. Biophys. Acta 1763:1669-1677. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

