Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis
- PMID: 17644729
- PMCID: PMC1955706
- DOI: 10.1105/tpc.106.045732
Functional analysis of the epidermal-specific MYB genes CAPRICE and WEREWOLF in Arabidopsis
Abstract
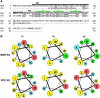
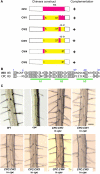
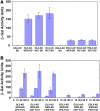
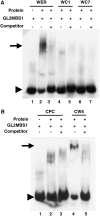
Epidermis cell differentiation in Arabidopsis thaliana is a model system for understanding the developmental end state of plant cells. Two types of MYB transcription factors, R2R3-MYB and R3-MYB, are involved in cell fate determination. To examine the molecular basis of this process, we analyzed the functional relationship of the R2R3-type MYB gene WEREWOLF (WER) and the R3-type MYB gene CAPRICE (CPC). Chimeric constructs made from the R3 MYB regions of WER and CPC used in reciprocal complementation experiments showed that the CPC R3 region cannot functionally substitute for the WER R3 region in the differentiation of hairless cells. However, WER R3 can substantially substitute for CPC R3. There are no differences in yeast interaction assays of WER or WER chimera proteins with GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3). CPC and CPC chimera proteins also have similar activity in preventing GL3 WER and EGL3 WER interactions. Furthermore, we showed by gel mobility shift assays that WER chimera proteins do not bind to the GL2 promoter region. However, a CPC chimera protein, which harbors the WER R3 motif, still binds to the GL2 promoter region.
Figures




References
-
- Andersson, K.B., Berge, T., Matre, V., and Gabrielsen, O.S. (1999). Sequence selectivity of c-Myb in vivo. Resolution of a DNA target specificity paradox. J. Biol. Chem. 274 21986–21994. - PubMed
-
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
-
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316 1194–1199.
-
- Bernhardt, C., Lee, M.M., Gonzalez, A., Zhang, F., Lloyd, A., and Schiefelbein, J. (2003). The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130 6431–6439. - PubMed
-
- Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A., and Schiefelbein, J. (2005). The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132 291–298. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

