PolyADP-ribosylation and cancer
- PMID: 17645773
- PMCID: PMC11159171
- DOI: 10.1111/j.1349-7006.2007.00567.x
PolyADP-ribosylation and cancer
Abstract
The polyADP-ribosylation reaction results in a unique post-translational modification involved in various cellular processes and conditions, including DNA repair, transcriptional control, genomic stability, cell death and transformation. The existence of 17 members of the poly(ADP-ribose) polymerase (PARP) family has so far been documented, with overlapping functional consequences. PARP-1 is known to be involved in DNA base excision repair and this explains the susceptibility spectrum of PARP-1 knockout animals to genotoxic carcinogens. The fact that centrosome amplification is induced by a non-genotoxic inhibitor of PARP and in PARP-1 knockout mouse cells, is in line with aneuploidy, which is frequent in cancers. Genetically engineered animal models have revealed that PARP-1 and VPARP impact carcinogenesis. Furthermore, accumulating experimental evidence supports the utility of PARP and PARG inhibitors in cancer therapy and several clinical trials are now ongoing. Increasing NAD(+) levels by pharmacological supplementation with niacin has also been found to exert preventive effects against cancer. In the present review, recent research progress on polyADP-ribosylation related to neoplasia is summarized and discussed.
Figures
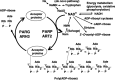



References
-
- Sugimura T. Poly (adenosine diphosphate ribose). Prog Nucl Acid Res Mol Biol 1973; 13: 127–51. - PubMed
-
- Miwa M, Kanai M, Uchida M, Uchida K, Hanai S. Roles of poly(ADP‐ribose) metabolism in the regulation of centrosome duplication and in the maintenance of neuronal integrity. In: Buerkle A, ed. Poly(ADP‐Ribosyl)ation. Georgetown, Texas: Landes Bioscience, 2006: 51–60
-
- Althaus FR, Richter C. ADP‐ribosylation of proteins. Enzymology and biological significance. Mol Biol Biochem Biophys 1987; 37: 1–237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

