Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations
- PMID: 17645794
- PMCID: PMC1971073
- DOI: 10.1186/1471-2164-8-245
Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations
Abstract
Background: Folate synthesis and salvage pathways are relatively well known from classical biochemistry and genetics but they have not been subjected to comparative genomic analysis. The availability of genome sequences from hundreds of diverse bacteria, and from Arabidopsis thaliana, enabled such an analysis using the SEED database and its tools. This study reports the results of the analysis and integrates them with new and existing experimental data.
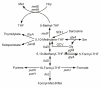
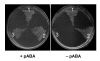
Results: Based on sequence similarity and the clustering, fusion, and phylogenetic distribution of genes, several functional predictions emerged from this analysis. For bacteria, these included the existence of novel GTP cyclohydrolase I and folylpolyglutamate synthase gene families, and of a trifunctional p-aminobenzoate synthesis gene. For plants and bacteria, the predictions comprised the identities of a 'missing' folate synthesis gene (folQ) and of a folate transporter, and the absence from plants of a folate salvage enzyme. Genetic and biochemical tests bore out these predictions.
Conclusion: For bacteria, these results demonstrate that much can be learnt from comparative genomics, even for well-explored primary metabolic pathways. For plants, the findings particularly illustrate the potential for rapid functional assignment of unknown genes that have prokaryotic homologs, by analyzing which genes are associated with the latter. More generally, our data indicate how combined genomic analysis of both plants and prokaryotes can be more powerful than isolated examination of either group alone.
Figures






Similar articles
-
Enhancing pterin and para-aminobenzoate content is not sufficient to successfully biofortify potato tubers and Arabidopsis thaliana plants with folate.J Exp Bot. 2013 Sep;64(12):3899-909. doi: 10.1093/jxb/ert224. J Exp Bot. 2013. PMID: 23956417
-
Finding novel metabolic genes through plant-prokaryote phylogenomics.Trends Microbiol. 2007 Dec;15(12):563-70. doi: 10.1016/j.tim.2007.10.008. Epub 2007 Nov 9. Trends Microbiol. 2007. PMID: 17997099 Review.
-
Proteins of Unknown Biochemical Function: A Persistent Problem and a Roadmap to Help Overcome It.Plant Physiol. 2015 Nov;169(3):1436-42. doi: 10.1104/pp.15.00959. Epub 2015 Aug 12. Plant Physiol. 2015. PMID: 26269542 Free PMC article.
-
Synthesis and turnover of folates in plants.Curr Opin Plant Biol. 2002 Jun;5(3):244-9. doi: 10.1016/s1369-5266(02)00249-2. Curr Opin Plant Biol. 2002. PMID: 11960743 Review.
-
Improved folate accumulation in genetically modified maize and wheat.J Exp Bot. 2019 Mar 11;70(5):1539-1551. doi: 10.1093/jxb/ery453. J Exp Bot. 2019. PMID: 30753561 Free PMC article.
Cited by
-
Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis.Cell. 2015 Apr 9;161(2):264-76. doi: 10.1016/j.cell.2015.02.047. Cell. 2015. PMID: 25860609 Free PMC article.
-
Comparative genome analysis of Pediococcus damnosus LMG 28219, a strain well-adapted to the beer environment.BMC Genomics. 2015 Apr 3;16(1):267. doi: 10.1186/s12864-015-1438-z. BMC Genomics. 2015. PMID: 25880122 Free PMC article.
-
Moonlighting glutamate formiminotransferases can functionally replace 5-formyltetrahydrofolate cycloligase.J Biol Chem. 2010 Dec 31;285(53):41557-66. doi: 10.1074/jbc.M110.190504. Epub 2010 Oct 15. J Biol Chem. 2010. PMID: 20952389 Free PMC article.
-
Identification of Efflux Substrates Using a Riboswitch-Based Reporter in Pseudomonas aeruginosa.mSphere. 2023 Apr 20;8(2):e0006923. doi: 10.1128/msphere.00069-23. Epub 2023 Mar 22. mSphere. 2023. PMID: 36946743 Free PMC article.
-
Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host.Proc Biol Sci. 2014 Dec 7;281(1796):20141838. doi: 10.1098/rspb.2014.1838. Proc Biol Sci. 2014. PMID: 25339726 Free PMC article.
References
-
- Matthews RG. One-carbon metabolism. In: Neidhardt FC, Curtiss R 3rd, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Second. Vol. 1. Washington DC, ASM Press; 1996. pp. 600–611.
-
- Huennekens FM, Vitols KS, Pope LE, Fan J. Membrane transport of folate compounds. J Nutr Sci Vitaminol (Tokyo) 1992:52–57. - PubMed
-
- Green JC, Nichols BP, Matthews RG. Folate biosynthesis, reduction, and polyglutamylation. In: Neidhardt FC, Curtiss R 3rd, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. Second. Vol. 1. Washington DC, ASM Press; 1996. pp. 665–673.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

