Global analysis of patterns of gene expression during Drosophila embryogenesis
- PMID: 17645804
- PMCID: PMC2323238
- DOI: 10.1186/gb-2007-8-7-r145
Global analysis of patterns of gene expression during Drosophila embryogenesis
Abstract
Background: Cell and tissue specific gene expression is a defining feature of embryonic development in multi-cellular organisms. However, the range of gene expression patterns, the extent of the correlation of expression with function, and the classes of genes whose spatial expression are tightly regulated have been unclear due to the lack of an unbiased, genome-wide survey of gene expression patterns.
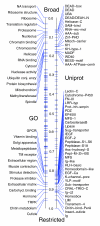
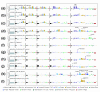
Results: We determined and documented embryonic expression patterns for 6,003 (44%) of the 13,659 protein-coding genes identified in the Drosophila melanogaster genome with over 70,000 images and controlled vocabulary annotations. Individual expression patterns are extraordinarily diverse, but by supplementing qualitative in situ hybridization data with quantitative microarray time-course data using a hybrid clustering strategy, we identify groups of genes with similar expression. Of 4,496 genes with detectable expression in the embryo, 2,549 (57%) fall into 10 clusters representing broad expression patterns. The remaining 1,947 (43%) genes fall into 29 clusters representing restricted expression, 20% patterned as early as blastoderm, with the majority restricted to differentiated cell types, such as epithelia, nervous system, or muscle. We investigate the relationship between expression clusters and known molecular and cellular-physiological functions.
Conclusion: Nearly 60% of the genes with detectable expression exhibit broad patterns reflecting quantitative rather than qualitative differences between tissues. The other 40% show tissue-restricted expression; the expression patterns of over 1,500 of these genes are documented here for the first time. Within each of these categories, we identified clusters of genes associated with particular cellular and developmental functions.
Figures







References
-
- Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat Genet. 1999;21:33–37. - PubMed
-
- Jones KW, Robertson FW. Localisation of reiterated nucleotide sequences in Drosophila and mouse by in situ hybridisation of complementary RNA. Chromosoma. 1970;31:331–345. - PubMed
-
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. - PubMed
-
- Arbeitman MN, Furlong EE, Imam F, Johnson E, Null BH, Baker BS, Krasnow MA, Scott MP, Davis RW, White KP. Gene expression during the life cycle of Drosophila melanogaster. Science. 2002;297:2270–2275. - PubMed
-
- Furlong EE, Andersen EC, Null B, White KP, Scott MP. Patterns of gene expression during Drosophila mesoderm development. Science. 2001;293:1629–1633. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

