Numerical modelling of label-structured cell population growth using CFSE distribution data
- PMID: 17650320
- PMCID: PMC1950697
- DOI: 10.1186/1742-4682-4-26
Numerical modelling of label-structured cell population growth using CFSE distribution data
Abstract
Background: The flow cytometry analysis of CFSE-labelled cells is currently one of the most informative experimental techniques for studying cell proliferation in immunology. The quantitative interpretation and understanding of such heterogenous cell population data requires the development of distributed parameter mathematical models and computational techniques for data assimilation.
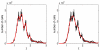
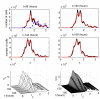
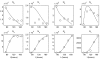
Methods and results: The mathematical modelling of label-structured cell population dynamics leads to a hyperbolic partial differential equation in one space variable. The model contains fundamental parameters of cell turnover and label dilution that need to be estimated from the flow cytometry data on the kinetics of the CFSE label distribution. To this end a maximum likelihood approach is used. The Lax-Wendroff method is used to solve the corresponding initial-boundary value problem for the model equation. By fitting two original experimental data sets with the model we show its biological consistency and potential for quantitative characterization of the cell division and death rates, treated as continuous functions of the CFSE expression level.
Conclusion: Once the initial distribution of the proliferating cell population with respect to the CFSE intensity is given, the distributed parameter modelling allows one to work directly with the histograms of the CFSE fluorescence without the need to specify the marker ranges. The label-structured model and the elaborated computational approach establish a quantitative basis for more informative interpretation of the flow cytometry CFSE systems.
Figures







Similar articles
-
Distributed parameter identification for a label-structured cell population dynamics model using CFSE histogram time-series data.J Math Biol. 2009 Nov;59(5):581-603. doi: 10.1007/s00285-008-0244-5. Epub 2008 Dec 19. J Math Biol. 2009. PMID: 19096849
-
A division-dependent compartmental model for computing cell numbers in CFSE-based lymphocyte proliferation assays.Math Biosci Eng. 2012 Oct;9(4):699-736. doi: 10.3934/mbe.2012.9.699. Math Biosci Eng. 2012. PMID: 23311419
-
Computational analysis of CFSE proliferation assay.J Math Biol. 2007 Jan;54(1):57-89. doi: 10.1007/s00285-006-0046-6. Epub 2006 Nov 9. J Math Biol. 2007. PMID: 17093999
-
Flow cytometric analysis of cell division by dilution of CFSE and related dyes.Curr Protoc Cytom. 2013;Chapter 9:9.11.1-9.11.12. doi: 10.1002/0471142956.cy0911s64. Curr Protoc Cytom. 2013. PMID: 23546777 Review.
-
Asymmetry of Cell Division in CFSE-Based Lymphocyte Proliferation Analysis.Front Immunol. 2013 Sep 2;4:264. doi: 10.3389/fimmu.2013.00264. Front Immunol. 2013. PMID: 24032033 Free PMC article. Review.
Cited by
-
Cyton2: A Model of Immune Cell Population Dynamics That Includes Familial Instructional Inheritance.Front Bioinform. 2021 Oct 26;1:723337. doi: 10.3389/fbinf.2021.723337. eCollection 2021. Front Bioinform. 2021. PMID: 36303793 Free PMC article.
-
Label Structured Cell Proliferation Models.Appl Math Lett. 2010 Dec 1;23(12):1412-1415. doi: 10.1016/j.aml.2010.07.009. Appl Math Lett. 2010. PMID: 20847901 Free PMC article.
-
Prediction of PD-L1 inhibition effects for HIV-infected individuals.PLoS Comput Biol. 2019 Nov 6;15(11):e1007401. doi: 10.1371/journal.pcbi.1007401. eCollection 2019 Nov. PLoS Comput Biol. 2019. PMID: 31693657 Free PMC article.
-
Mathematical models for CFSE labelled lymphocyte dynamics: asymmetry and time-lag in division.J Math Biol. 2014 Dec;69(6-7):1547-83. doi: 10.1007/s00285-013-0741-z. Epub 2013 Dec 13. J Math Biol. 2014. PMID: 24337680
-
Distributed parameter identification for a label-structured cell population dynamics model using CFSE histogram time-series data.J Math Biol. 2009 Nov;59(5):581-603. doi: 10.1007/s00285-008-0244-5. Epub 2008 Dec 19. J Math Biol. 2009. PMID: 19096849
References
-
- Srienc F. Cytometric data as the basis for rigorous models of cell population dynamics. J Biotechnol. 1999;71:233–238. doi: 10.1016/S0168-1656(99)00026-7. - DOI
-
- Lyons AB, Doherty KV. Flow cytometric analysis of cell division by dye dilution. Current Protocols in Cytometry. 2004. pp. 9.11.1–9.11.10. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

