Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori)
- PMID: 17651473
- PMCID: PMC1976426
- DOI: 10.1186/1471-213X-7-88
Characterization and expression patterns of let-7 microRNA in the silkworm (Bombyx mori)
Abstract
Background: lin-4 and let-7, the two founding members of heterochronic microRNA genes, are firstly confirmed in Caenorhabditis elegans to control the proper timing of developmental programs in a heterochronic pathway. let-7 has been thought to trigger the onset of adulthood across animal phyla. Ecdysone and Broad-Complex are required for the temporal expression of let-7 in Drosophila melanogaster. For a better understanding of the conservation and functions of let-7, we seek to explore how it is expressed in the silkworm (Bombyx mori).
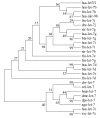
Results: One member of let-7 family has been identified in silkworm computationally and experimentally. All known members of this family share the same nucleotides at ten positions within the mature sequences. Sequence logo and phylogenetic tree show that they are not only conserved but diversify to some extent among some species. The bmo-let-7 was very lowly expressed in ova harvested from newborn unmated female adult and in individuals from the first molt to the early third instar, highly expressed after the third molt, and the most abundant expression was observed after mounting, particularly after pupation. The expression levels were higher at the end of each instar and at the beginning of each molt than at other periods, coinciding with the pulse of ecdysone and BR-C as a whole. Using cultured ovary cell line, BmN-SWU1, we examined the effect of altered ecdysone levels on bmo-let-7 expression. The expression was also detected in various tissues of day 3 of the fifth instar and of from day 7 of the fifth to pupa, suggesting a wide distributing pattern with various signal intensities.
Conclusion: bmo-let-7 is stage- and tissue-specifically expressed in the silkworm. Although no signals were detected during embryonic development and first larval instar stages, the expression of bmo-let-7 was observed from the first molt, suggesting that it might also function at early larval stage of the silkworm. The detailed expression profiles in the whole life cycle and cultured cell line of silkworm showed a clear association with ecdysone pulse and a variety of biological processes.
Figures










References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

