Fluorescent labeling of tRNAs for dynamics experiments
- PMID: 17652134
- PMCID: PMC1950756
- DOI: 10.1261/rna.475407
Fluorescent labeling of tRNAs for dynamics experiments
Abstract
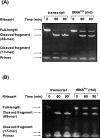
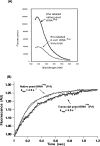
Transfer RNAs (tRNAs) are substrates for complex enzymes, such as aminoacyl-tRNA synthetases and ribosomes, and play an essential role in translation of genetic information into protein sequences. Here we describe a general method for labeling tRNAs with fluorescent dyes, so that the activities and dynamics of the labeled tRNAs can be directly monitored by fluorescence during the ribosomal decoding process. This method makes use of the previously reported fluorescent labeling of natural tRNAs at dihydrouridine (D) positions, but extends the previous method to synthetic tRNAs by preparing tRNA transcripts and introducing D residues into transcripts with the yeast enzyme Dus1p dihydrouridine synthase. Using the unmodified transcript of Escherichia coli tRNAPro as an example, which has U17 and U17a in the D loop, we show that Dus1p catalyzes conversion of one of these Us (mostly U17a) to D, and that the modified tRNA can be labeled with the fluorophores proflavin and rhodamine 110, with overall labeling yields comparable to those obtained with the native yeast tRNAPhe. Further, the transcript of yeast tRNAPhe, modified by Dus1p and labeled with proflavin, translocates on the ribosome at a rate similar to that of the proflavin-labeled native yeast tRNAPhe. These results demonstrate that synthetic tRNA transcripts, which may be designed to contain mutations not found in nature, can be labeled and studied. Such labeled tRNAs should have broad utility in research that involves studies of tRNA maturation, aminoacylation, and tRNA-ribosome interactions.
Figures
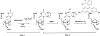


References
-
- Bishop, A.C., Xu, J., Johnson, R.C., Schimmel, P., de Crecy-Lagard, V. Identification of the tRNA–dihydrouridine synthase family. J. Biol. Chem. 2002;277:25090–25095. - PubMed
-
- Blanchard, S.C., Gonzalez, R.L., Kim, H.D., Chu, S., Puglisi, J.D. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004a;11:1008–1014. - PubMed
-
- Boonacker, E., Van Noorden, C. Enzyme cytochemical techniques for metabolic mapping in living cells with special reference to proteolysis. J. Histochem. Cytochem. 2001;49:1473–1486. - PubMed
-
- Cerutti, P., Miller, N. Selective reduction of yeast transfer ribonucleic acid with sodium borohydride. J. Mol. Biol. 1967;26:55–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
