Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family
- PMID: 17652178
- PMCID: PMC1935027
- DOI: 10.1101/gad.1561707
Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family
Abstract
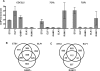
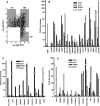
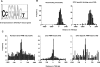
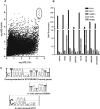
The conservation of in vitro DNA-binding properties within families of transcription factors presents a challenge for achieving in vivo specificity. To uncover the mechanisms regulating specificity within the ETS gene family, we have used chromatin immunoprecipitation coupled with genome-wide promoter microarrays to query the occupancy of three ETS proteins in a human T-cell line. Unexpectedly, redundant occupancy was frequently detected, while specific occupancy was less likely. Redundant binding correlated with housekeeping classes of genes, whereas specific binding examples represented more specialized genes. Bioinformatics approaches demonstrated that redundant binding correlated with consensus ETS-binding sequences near transcription start sites. In contrast, specific binding sites diverged dramatically from the consensus and were found further from transcription start sites. One route to specificity was found--a highly divergent binding site that facilitates ETS1 and RUNX1 cooperative DNA binding. The specific and redundant DNA-binding modes suggest two distinct roles for members of the ETS transcription factor family.
Figures

References
-
- Aerts S., Van Loo P., Thijs G., Moreau Y., De Moor B., Van Loo P., Thijs G., Moreau Y., De Moor B., Thijs G., Moreau Y., De Moor B., Moreau Y., De Moor B., De Moor B. Computational detection of cis-regulatory modules. Bioinformatics. 2003;19 (Suppl. 2):II5–II14. - PubMed
-
- Bailey T.L., Elkan C., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Altman R., et al., editors. Proceedings of the second international conference on intelligent systems for molecular biology. AAAI Press; Menlo Park, CA: 1994. pp. 28–36. - PubMed
-
- Batchelor A.H., Piper D.E., de la Brousse F.C., McKnight S.L., Wolberger C., Piper D.E., de la Brousse F.C., McKnight S.L., Wolberger C., de la Brousse F.C., McKnight S.L., Wolberger C., McKnight S.L., Wolberger C., Wolberger C. The structure of GABPα/β: An ETS domain–ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. - PubMed
-
- Beissbarth T., Speed T.P., Speed T.P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. - PubMed
-
- Bina M., Wyss P., Ren W., Szpankowski W., Thomas E., Randhawa R., Reddy S., John P.M., Pares-Matos E.I., Stein A., Wyss P., Ren W., Szpankowski W., Thomas E., Randhawa R., Reddy S., John P.M., Pares-Matos E.I., Stein A., Ren W., Szpankowski W., Thomas E., Randhawa R., Reddy S., John P.M., Pares-Matos E.I., Stein A., Szpankowski W., Thomas E., Randhawa R., Reddy S., John P.M., Pares-Matos E.I., Stein A., Thomas E., Randhawa R., Reddy S., John P.M., Pares-Matos E.I., Stein A., Randhawa R., Reddy S., John P.M., Pares-Matos E.I., Stein A., Reddy S., John P.M., Pares-Matos E.I., Stein A., John P.M., Pares-Matos E.I., Stein A., Pares-Matos E.I., Stein A., Stein A., et al. Exploring the characteristics of sequence elements in proximal promoters of human genes. Genomics. 2004;84:929–940. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
