Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae
- PMID: 17652322
- PMCID: PMC1976441
- DOI: 10.1093/nar/gkm442
Ionizing radiation and restriction enzymes induce microhomology-mediated illegitimate recombination in Saccharomyces cerevisiae
Abstract
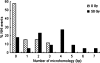
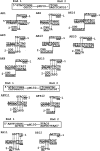
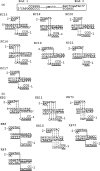
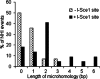
DNA double-strand breaks can be repaired by illegitimate recombination without extended sequence homology. A distinct mechanism namely microhomology-mediated recombination occurs between a few basepairs of homology that is associated with deletions. Ionizing radiation and restriction enzymes have been shown to increase the frequency of nonhomologous integration in yeast. However, the mechanism of such enhanced recombination events is not known. Here, we report that both ionizing radiation and restriction enzymes increase the frequency of microhomology-mediated integration. Irradiated yeast cells displayed 77% microhomology-mediated integration, compared to 27% in unirradiated cells. Radiation-induced integration exhibited lack of deletions at genomic insertion sites, implying that such events are likely to occur at undamaged sites. Restriction enzymes also enhanced integration events at random non-restriction sites via microhomology-mediated recombination. Furthermore, generation of a site-specific I-SceI-mediated double-strand break induces microhomology-mediated integration randomly throughout the genome. Taken together, these results suggest that double-strand breaks induce a genome-wide microhomology-mediated illegitimate recombination pathway that facilitates integration probably in trans at non-targeted sites and might be involved in generation of large deletions and other genomic rearrangements.
Figures


Similar articles
-
Rad1, rad10 and rad52 mutations reduce the increase of microhomology length during radiation-induced microhomology-mediated illegitimate recombination in saccharomyces cerevisiae.Radiat Res. 2009 Aug;172(2):141-51. doi: 10.1667/RR1675.1. Radiat Res. 2009. PMID: 19630519 Free PMC article.
-
Ionizing radiation induces microhomology-mediated end joining in trans in yeast and mammalian cells.Radiat Res. 2009 Apr;171(4):454-63. doi: 10.1667/RR1329.1. Radiat Res. 2009. PMID: 19397446 Free PMC article.
-
Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells.DNA Repair (Amst). 2007 Jun 1;6(6):781-8. doi: 10.1016/j.dnarep.2007.01.004. Epub 2007 Feb 12. DNA Repair (Amst). 2007. PMID: 17296333
-
Ionizing radiation and genetic risks XIV. Potential research directions in the post-genome era based on knowledge of repair of radiation-induced DNA double-strand breaks in mammalian somatic cells and the origin of deletions associated with human genomic disorders.Mutat Res. 2005 Oct 15;578(1-2):333-70. doi: 10.1016/j.mrfmmm.2005.06.020. Epub 2005 Aug 5. Mutat Res. 2005. PMID: 16084534 Review.
-
Multi-site-specific endonucleases and the initiation of homologous genetic recombination in yeast.Adv Biophys. 1995;31:77-91. doi: 10.1016/0065-227x(95)99384-2. Adv Biophys. 1995. PMID: 7625280 Review.
Cited by
-
Aerospace Technology Improves Fermentation Potential of Microorganisms.Front Microbiol. 2022 Apr 29;13:896556. doi: 10.3389/fmicb.2022.896556. eCollection 2022. Front Microbiol. 2022. PMID: 35572688 Free PMC article. Review.
-
Mitochondrial genome plasticity of mammalian species.BMC Genomics. 2024 Mar 14;25(1):278. doi: 10.1186/s12864-024-10201-9. BMC Genomics. 2024. PMID: 38486136 Free PMC article.
-
Requirement of POL3 and POL4 on non-homologous and microhomology-mediated end joining in rad50/xrs2 mutants of Saccharomyces cerevisiae.Mutagenesis. 2015 Nov;30(6):841-9. doi: 10.1093/mutage/gev046. Epub 2015 Jun 29. Mutagenesis. 2015. PMID: 26122113 Free PMC article.
-
MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings.Trends Genet. 2008 Nov;24(11):529-38. doi: 10.1016/j.tig.2008.08.007. Epub 2008 Sep 21. Trends Genet. 2008. PMID: 18809224 Free PMC article. Review.
-
Rad1, rad10 and rad52 mutations reduce the increase of microhomology length during radiation-induced microhomology-mediated illegitimate recombination in saccharomyces cerevisiae.Radiat Res. 2009 Aug;172(2):141-51. doi: 10.1667/RR1675.1. Radiat Res. 2009. PMID: 19630519 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

