Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations
- PMID: 17653265
- PMCID: PMC1919426
- DOI: 10.1371/journal.pone.0000638
Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations
Abstract
Background: The success of antiretroviral therapy is known to be compromised by drug-resistant HIV-1 at frequencies detectable by conventional bulk sequencing. Currently, there is a need to assess the clinical consequences of low-frequency drug resistant variants occurring below the detection limit of conventional genotyping. Sensitive detection of drug-resistant subpopulations, however, requires simple and practical methods for routine testing.
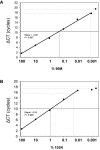
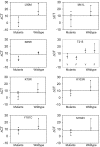
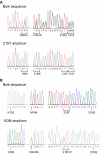
Methodology: We developed highly-sensitive and simple real-time PCR assays for nine key drug resistance mutations and show that these tests overcome substantial sequence heterogeneity in HIV-1 clinical specimens. We specifically used early wildtype virus samples from the pre-antiretroviral drug era to measure background reactivity and were able to define highly-specific screening cut-offs that are up to 67-fold more sensitive than conventional genotyping. We also demonstrate that sequencing the mutation-specific PCR products provided a direct and novel strategy to further detect and link associated resistance mutations, allowing easy identification of multi-drug-resistant variants. Resistance mutation associations revealed in mutation-specific amplicon sequences were verified by clonal sequencing.
Significance: Combined, sensitive real-time PCR testing and mutation-specific amplicon sequencing provides a powerful and simple approach that allows for improved detection and evaluation of HIV-1 drug resistance mutations.
Conflict of interest statement
Figures
Similar articles
-
A sensitive in-house RT-PCR genotyping system for combined detection of plasma HIV-1 and assessment of drug resistance.J Virol Methods. 2006 May;133(2):137-45. doi: 10.1016/j.jviromet.2005.11.004. Epub 2005 Dec 20. J Virol Methods. 2006. PMID: 16375980
-
Study of the impact of HIV genotypic drug resistance testing on therapy efficacy.Verh K Acad Geneeskd Belg. 2001;63(5):447-73. Verh K Acad Geneeskd Belg. 2001. PMID: 11813503 Review.
-
Genotypic resistance tests for the management of the HIV-infected patient with non-B viral isolates.Scand J Infect Dis Suppl. 2003;106:75-8. Scand J Infect Dis Suppl. 2003. PMID: 15000590 Review.
-
Evaluation of minority populations of HIV type-1 with K103N and M184V drug resistance mutations among children in Argentina.Antivir Ther. 2009;14(8):1175-81. doi: 10.3851/IMP1461. Antivir Ther. 2009. PMID: 20032547
-
Evaluation of an in-house genotyping resistance test for HIV-1 drug resistance interpretation and genotyping.J Clin Virol. 2007 Jun;39(2):125-31. doi: 10.1016/j.jcv.2007.03.008. Epub 2007 Apr 20. J Clin Virol. 2007. PMID: 17449318
Cited by
-
Basis for early and preferential selection of the E138K mutation in HIV-1 reverse transcriptase.Antimicrob Agents Chemother. 2013 Oct;57(10):4681-8. doi: 10.1128/AAC.01029-13. Epub 2013 Jul 15. Antimicrob Agents Chemother. 2013. PMID: 23856772 Free PMC article.
-
Factors limiting the transmission of HIV mutations conferring drug resistance: fitness costs and genetic bottlenecks.Sci Rep. 2012;2:320. doi: 10.1038/srep00320. Epub 2012 Mar 19. Sci Rep. 2012. PMID: 22432052 Free PMC article.
-
Dual Simian Foamy Virus/Human Immunodeficiency Virus Type 1 Infections in Persons from Côte d'Ivoire.PLoS One. 2016 Jun 16;11(6):e0157709. doi: 10.1371/journal.pone.0157709. eCollection 2016. PLoS One. 2016. PMID: 27310836 Free PMC article.
-
Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naïve populations and associate with reduced treatment efficacy.PLoS Med. 2008 Jul 29;5(7):e158. doi: 10.1371/journal.pmed.0050158. PLoS Med. 2008. PMID: 18666824 Free PMC article.
-
Characterization of the Drug Resistance Profiles of Patients Infected with CRF07_BC Using Phenotypic Assay and Ultra-Deep Pyrosequencing.PLoS One. 2017 Jan 20;12(1):e0170420. doi: 10.1371/journal.pone.0170420. eCollection 2017. PLoS One. 2017. PMID: 28107423 Free PMC article.
References
-
- US Department of Health and Human Services. 2006. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. . Available: http://AIDSinfo.nih.gov.
-
- Johnson JA, Li J-F, Morris L, Martinson N, Gray G, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. - PubMed
-
- Flys T, Nissley DV, Claasen CW, Jones D, Shi C, et al. Sensitive drug resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005;192:24–29. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

