Knockdown of the bovine prion gene PRNP by RNA interference (RNAi) technology
- PMID: 17655742
- PMCID: PMC1976095
- DOI: 10.1186/1472-6750-7-44
Knockdown of the bovine prion gene PRNP by RNA interference (RNAi) technology
Abstract
Background: Since prion gene-knockout mice do not contract prion diseases and animals in which production of prion protein (PrP) is reduced by half are resistant to the disease, we hypothesized that bovine animals with reduced PrP would be tolerant to BSE. Hence, attempts were made to produce bovine PRNP (bPRNP) that could be knocked down by RNA interference (RNAi) technology. Before an in vivo study, optimal conditions for knocking down bPRNP were determined in cultured mammalian cell systems. Factors examined included siRNA (short interfering RNA) expression plasmid vectors, target sites of PRNP, and lengths of siRNAs.
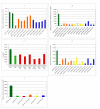
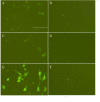
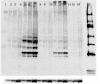
Results: Four siRNA expression plasmid vectors were used: three harboring different cloning sites were driven by the human U6 promoter (hU6), and one by the human tRNAVal promoter. Six target sites of bovine PRNP were designed using an algorithm. From 1 (22 mer) to 9 (19, 20, 21, 22, 23, 24, 25, 27, and 29 mer) siRNA expression vectors were constructed for each target site. As targets of siRNA, the entire bPRNP coding sequence was connected to the reporter gene of the fluorescent EGFP, or of firefly luciferase or Renilla luciferase. Target plasmid DNA was co-transfected with siRNA expression vector DNA into HeLaS3 cells, and fluorescence or luminescence was measured. The activities of siRNAs varied widely depending on the target sites, length of the siRNAs, and vectors used. Longer siRNAs were less effective, and 19 mer or 21 mer was generally optimal. Although 21 mer GGGGAGAACTTCACCGAAACT expressed by a hU6-driven plasmid with a Bsp MI cloning site was best under the present experimental conditions, the corresponding tRNA promoter-driven plasmid was almost equally useful. The effectiveness of this siRNA was confirmed by immunostaining and Western blotting.
Conclusion: Four siRNA expression plasmid vectors, six target sites of bPRNP, and various lengths of siRNAs from 19 mer to 29 mer were examined to establish optimal conditions for knocking down of bPRNP in vitro. The most effective siRNA so far tested was 21 mer GGGGAGAACTTCACCGAAACT driven either by a hU6 or tRNA promoter, a finding that provides a basis for further studies in vivo.
Figures
Similar articles
-
Knockdown of the prion gene expression by RNA interference in bovine fibroblast cells.Mol Biol Rep. 2010 Oct;37(7):3193-8. doi: 10.1007/s11033-009-9900-0. Epub 2009 Oct 11. Mol Biol Rep. 2010. PMID: 19821149
-
Combination of the somatic cell nuclear transfer method and RNAi technology for the production of a prion gene-knockdown calf using plasmid vectors harboring the U6 or tRNA promoter.Prion. 2011 Jan-Mar;5(1):39-46. doi: 10.4161/pri.5.1.14075. Epub 2011 Jan 1. Prion. 2011. PMID: 21084838 Free PMC article.
-
Mouse neuronal cells expressing exogenous bovine PRNP and simultaneous downregulation of endogenous mouse PRNP using siRNAs.Prion. 2010 Jan-Mar;4(1):32-7. doi: 10.4161/pri.4.1.11218. Epub 2010 Jan 15. Prion. 2010. PMID: 20215868 Free PMC article.
-
Small RNA: can RNA interference be exploited for therapy?Lancet. 2003 Oct 25;362(9393):1401-3. doi: 10.1016/S0140-6736(03)14637-5. Lancet. 2003. PMID: 14585643 Review.
-
Therapeutic potential of siRNA-mediated transcriptional gene silencing.Biotechniques. 2006 Apr;Suppl:7-13. doi: 10.2144/000112166. Biotechniques. 2006. PMID: 16629382 Review.
Cited by
-
A new method, using cis-regulatory control, for blocking embryonic gene expression.Dev Biol. 2008 Jun 15;318(2):360-5. doi: 10.1016/j.ydbio.2008.02.056. Epub 2008 Mar 14. Dev Biol. 2008. PMID: 18423438 Free PMC article.
-
Efficient PRNP deletion in bovine genome using gene-editing technologies in bovine cells.Prion. 2015;9(4):278-91. doi: 10.1080/19336896.2015.1071459. Prion. 2015. PMID: 26217959 Free PMC article.
-
Establishment and characterization of Prnp knockdown neuroblastoma cells using dual microRNA-mediated RNA interference.Prion. 2011 Apr-Jun;5(2):93-102. doi: 10.4161/pri.5.2.15621. Epub 2011 Apr 1. Prion. 2011. PMID: 21494092 Free PMC article.
-
Knockdown of the prion gene expression by RNA interference in bovine fibroblast cells.Mol Biol Rep. 2010 Oct;37(7):3193-8. doi: 10.1007/s11033-009-9900-0. Epub 2009 Oct 11. Mol Biol Rep. 2010. PMID: 19821149
-
Combination of the somatic cell nuclear transfer method and RNAi technology for the production of a prion gene-knockdown calf using plasmid vectors harboring the U6 or tRNA promoter.Prion. 2011 Jan-Mar;5(1):39-46. doi: 10.4161/pri.5.1.14075. Epub 2011 Jan 1. Prion. 2011. PMID: 21084838 Free PMC article.
References
-
- Kempster S, Collins ME, Deacon R, Edington N. Impaired motor coordination on static rods in BSE-infected mice. Behav Brain Res. 2004;154:291–295. - PubMed
-
- Sakaguchi S, Katamine S, Nishida N, Moriuchi R, Shigematsu K, Sugimoto T, Nakatani A, Kataoka Y, Houtani T, Shirabe S, Okada H, Hasegawa S, Miyamoto T, Noda T. Loss of cerebellar Purkinje cells in aged mice homozygous for a disrupted PrP gene. Nature. 1996;380:528–31. doi: 10.1038/380528a0. - DOI - PubMed
-
- Nico PB, de-Paris F, Vinade ER, Amaral OB, Rockenbach I, Soares BL, Guarnieri R, Wichert-Ana L, Calvo F, Walz R, Izquierdo I, Sakamoto AC, Brentani R, Martins VR, Bianchin MM. Altered behavioural response to acute stress in mice lacking cellular prion protein. Behav Brain Res. 2005;162:173–181. doi: 10.1016/j.bbr.2005.02.003. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

