Reactivation of methionine synthase from Thermotoga maritima (TM0268) requires the downstream gene product TM0269
- PMID: 17656578
- PMCID: PMC2203375
- DOI: 10.1110/ps.072936307
Reactivation of methionine synthase from Thermotoga maritima (TM0268) requires the downstream gene product TM0269
Abstract
The crystal structure of the Thermotoga maritima gene product TM0269, determined as part of genome-wide structural coverage of T. maritima by the Joint Center for Structural Genomics, revealed structural homology with the fourth module of the cobalamin-dependent methionine synthase (MetH) from Escherichia coli, despite the lack of significant sequence homology. The gene specifying TM0269 lies in close proximity to another gene, TM0268, which shows sequence homology with the first three modules of E. coli MetH. The fourth module of E. coli MetH is required for reductive remethylation of the cob(II)alamin form of the cofactor and binds the methyl donor for this reactivation, S-adenosylmethionine (AdoMet). Measurements of the rates of methionine formation in the presence and absence of TM0269 and AdoMet demonstrate that both TM0269 and AdoMet are required for reactivation of the inactive cob(II)alamin form of TM0268. These activity measurements confirm the structure-based assignment of the function of the TM0269 gene product. In the presence of TM0269, AdoMet, and reductants, the measured activity of T. maritima MetH is maximal near 80 degrees C, where the specific activity of the purified protein is approximately 15% of that of E. coli methionine synthase (MetH) at 37 degrees C. Comparisons of the structures and sequences of TM0269 and the reactivation domain of E. coli MetH suggest that AdoMet may be bound somewhat differently by the homologous proteins. However, the conformation of a hairpin that is critical for cobalamin binding in E. coli MetH, which constitutes an essential structural element, is retained in the T. maritima reactivation protein despite striking divergence of the sequences.
Figures

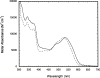
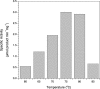
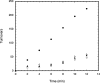
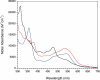

Similar articles
-
The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12.Structure. 1996 Nov 15;4(11):1263-75. doi: 10.1016/s0969-2126(96)00135-9. Structure. 1996. PMID: 8939751
-
Methionine synthase exists in two distinct conformations that differ in reactivity toward methyltetrahydrofolate, adenosylmethionine, and flavodoxin.Biochemistry. 1998 Apr 21;37(16):5372-82. doi: 10.1021/bi9730893. Biochemistry. 1998. PMID: 9548919
-
The mechanism of adenosylmethionine-dependent activation of methionine synthase: a rapid kinetic analysis of intermediates in reductive methylation of Cob(II)alamin enzyme.Biochemistry. 1998 Sep 8;37(36):12649-58. doi: 10.1021/bi9808565. Biochemistry. 1998. PMID: 9730838
-
Cobalamin-dependent methionine synthase: the structure of a methylcobalamin-binding fragment and implications for other B12-dependent enzymes.Curr Opin Struct Biol. 1994 Dec;4(6):919-29. doi: 10.1016/0959-440x(94)90275-5. Curr Opin Struct Biol. 1994. PMID: 7712296 Review.
-
Binding site revealed of nature's most beautiful cofactor.Science. 1994 Dec 9;266(5191):1663-4. doi: 10.1126/science.7992049. Science. 1994. PMID: 7992049 Review. No abstract available.
Cited by
-
Four families of folate-independent methionine synthases.PLoS Genet. 2021 Feb 3;17(2):e1009342. doi: 10.1371/journal.pgen.1009342. eCollection 2021 Feb. PLoS Genet. 2021. PMID: 33534785 Free PMC article.
-
Conformational switching and flexibility in cobalamin-dependent methionine synthase studied by small-angle X-ray scattering and cryo-electron microscopy.bioRxiv [Preprint]. 2023 Feb 12:2023.02.11.528079. doi: 10.1101/2023.02.11.528079. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2023 Jun 27;120(26):e2302531120. doi: 10.1073/pnas.2302531120. PMID: 36798380 Free PMC article. Updated. Preprint.
-
Conformational switching and flexibility in cobalamin-dependent methionine synthase studied by small-angle X-ray scattering and cryoelectron microscopy.Proc Natl Acad Sci U S A. 2023 Jun 27;120(26):e2302531120. doi: 10.1073/pnas.2302531120. Epub 2023 Jun 20. Proc Natl Acad Sci U S A. 2023. PMID: 37339208 Free PMC article.
-
The structural basis of protein conformational switching revealed by experimental and AlphaFold2 analyses.Proc Natl Acad Sci U S A. 2023 Jul 25;120(30):e2309689120. doi: 10.1073/pnas.2309689120. Epub 2023 Jul 13. Proc Natl Acad Sci U S A. 2023. PMID: 37440570 Free PMC article. No abstract available.
-
The ether-cleaving methyltransferase system of the strict anaerobe Acetobacterium dehalogenans: analysis and expression of the encoding genes.J Bacteriol. 2009 Jan;191(2):588-99. doi: 10.1128/JB.01104-08. Epub 2008 Nov 14. J Bacteriol. 2009. PMID: 19011025 Free PMC article.
References
-
- Bandarian V. and Matthews, R.G. 2001. Quantitation of rate enhancements attained by the binding of cobalamin to methionine synthase. Biochemistry 40: 5056–5064. - PubMed
-
- Bandarian V., Pattridge, K.A., Lennon, B.W., Huddler, D.P., Matthews, R.G., and Ludwig, M.L. 2002. Domain alternation switches B12-dependent methionine synthase to the activation conformation. Nat. Struct. Biol. 9: 53–56. - PubMed
-
- Banerjee R.V., Johnston, N.L., Sobeski, J.K., Datta, P., and Matthews, R.G. 1989. Cloning and sequence analysis of the Escherichia coli metH gene encoding cobalamin-dependent methionine synthase and isolation of a tryptic fragment containing the cobalamin-binding domain. J. Biol. Chem. 264: 13888–13895. - PubMed
-
- Banerjee R.V., Harder, S.F., Ragsdale, S.W., and Matthews, R.G. 1990. Mechanism of reductive activation of cobalamin-dependent methionine synthase: An electron paramagnetic resonance spectroelectrochemical study. Biochemistry 29: 1129–1133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

