Optimizing a multicolor immunophenotyping assay
- PMID: 17658403
- PMCID: PMC2034273
- DOI: 10.1016/j.cll.2007.05.002
Optimizing a multicolor immunophenotyping assay
Abstract
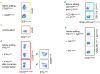
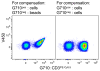
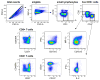
Flow cytometry-based immunophenotyping assays have become increasingly multiparametric, concomitantly analyzing multiple cellular parameters. To maximize the quality of the information obtained, antibody conjugate panels need to be developed with care, including requisite controls at every step. Such an optimization procedure for multicolor immunophenotyping assays is time consuming, but the value of having a reliable antibody conjugate panel that provides for sensitive detection of all molecules of interest justifies this time investment. This article outlines important considerations and procedures to undertake for the successful design and development of multicolor flow cytometry panels.
Figures




References
-
- Heeney JL, Plotkin SA. Immunological correlates of protection from HIV infection and disease. Nat Immunol. 2006 Dec;7(12):1281–1284. - PubMed
-
- Roederer M, Brenchley JM, Betts MR, De Rosa SC. Flow cytometric analysis of vaccine responses: how many colors are enough? Clin Immunol. 2004 Mar;110(3):199–205. - PubMed
-
- Perfetto SP, Roederer M. Increased immunofluorescence sensitivity using 532 nm laser excitation. Cytometry A. 2007 Jan 2; - PubMed
-
- Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004 Aug;4(8):648–655. - PubMed
-
- Perfetto SP, Chattopadhyay PK, Lamoreaux L, et al. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. 2006 Jun 30;313(1–2):199–208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

