Expanding the repertoire of an ERK2 recruitment site: cysteine footprinting identifies the D-recruitment site as a mediator of Ets-1 binding
- PMID: 17658891
- PMCID: PMC2897722
- DOI: 10.1021/bi7002058
Expanding the repertoire of an ERK2 recruitment site: cysteine footprinting identifies the D-recruitment site as a mediator of Ets-1 binding
Abstract
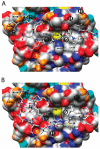
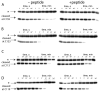
Many substrates of ERK2 contain a D-site, a sequence recognized by ERK2 that is used to promote catalysis. Despite lacking a canonical D-site, the substrate Ets-1 is displaced from ERK2 by peptides containing one. This suggests that Ets-1 may contain a novel or cryptic D-site. To investigate this possibility a protein footprinting strategy was developed to elucidate ERK2-ligand interactions. Using this approach, single cysteine reporters were placed in the D-recruitment site (DRS) of ERK2 and the resulting ERK2 proteins subjected to alkylation by iodoacetamide. The ability of residues 1-138 of Ets-1 to protect the cysteines from alkylation was determined. The pattern of protection observed is consistent with Ets-1 occupying a hydrophobic binding site within the DRS of ERK2. Significantly, a peptide derived from the D-site of Elk-1, which is known to bind the DRS, exhibits a similar pattern of cysteine protection. This analysis expands the repertoire of the DRS on ERK2 and suggests that other targeting sequences remain to be identified. Furthermore, cysteine-footprinting is presented as a useful way to interrogate protein-ligand interactions at the resolution of a single amino acid.
Figures
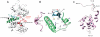





Similar articles
-
A model of a MAPK•substrate complex in an active conformation: a computational and experimental approach.PLoS One. 2011 Apr 11;6(4):e18594. doi: 10.1371/journal.pone.0018594. PLoS One. 2011. PMID: 21494553 Free PMC article.
-
The anti-apoptotic protein PEA-15 is a tight binding inhibitor of ERK1 and ERK2, which blocks docking interactions at the D-recruitment site.Biochemistry. 2007 Aug 14;46(32):9187-98. doi: 10.1021/bi700206u. Epub 2007 Jul 21. Biochemistry. 2007. PMID: 17658892
-
Properties and regulation of a transiently assembled ERK2.Ets-1 signaling complex.Biochemistry. 2006 Nov 21;45(46):13719-33. doi: 10.1021/bi0610451. Biochemistry. 2006. PMID: 17105191
-
A kinetic approach towards understanding substrate interactions and the catalytic mechanism of the serine/threonine protein kinase ERK2: identifying a potential regulatory role for divalent magnesium.Biochim Biophys Acta. 2004 Mar 11;1697(1-2):81-7. doi: 10.1016/j.bbapap.2003.11.015. Biochim Biophys Acta. 2004. PMID: 15023352 Review.
-
Quantifying ERK2-protein interactions by fluorescence anisotropy: PEA-15 inhibits ERK2 by blocking the binding of DEJL domains.Biochim Biophys Acta. 2005 Dec 30;1754(1-2):316-23. doi: 10.1016/j.bbapap.2005.11.002. Epub 2005 Nov 16. Biochim Biophys Acta. 2005. PMID: 16324895 Review.
Cited by
-
A model of a MAPK•substrate complex in an active conformation: a computational and experimental approach.PLoS One. 2011 Apr 11;6(4):e18594. doi: 10.1371/journal.pone.0018594. PLoS One. 2011. PMID: 21494553 Free PMC article.
-
P45 forms a complex with FADD and promotes neuronal cell survival following spinal cord injury.PLoS One. 2013 Jul 23;8(7):e69286. doi: 10.1371/journal.pone.0069286. Print 2013. PLoS One. 2013. PMID: 23935974 Free PMC article.
-
Identification and biochemical characterization of small molecule inhibitors of ERK2 that target the D-recruitment site.Methods Enzymol. 2023;690:445-499. doi: 10.1016/bs.mie.2023.06.016. Epub 2023 Aug 2. Methods Enzymol. 2023. PMID: 37858538 Free PMC article.
-
Computational insights for the discovery of non-ATP competitive inhibitors of MAP kinases.Curr Pharm Des. 2012;18(9):1173-85. doi: 10.2174/138161212799436368. Curr Pharm Des. 2012. PMID: 22316156 Free PMC article. Review.
-
Using Caenorhabditis elegans as a model organism for evaluating extracellular signal-regulated kinase docking domain inhibitors.J Cell Commun Signal. 2008 Dec;2(3-4):81-92. doi: 10.1007/s12079-008-0034-2. Epub 2008 Dec 23. J Cell Commun Signal. 2008. PMID: 19105050 Free PMC article.
References
-
- Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. - PubMed
-
- Cobb MH, Robbins DJ, Boulton TG. ERKs, extracellular signal-regulated MAP-2 kinases. Curr. Opin. Cell Biol. 1991 Dec;3:1025–1032. - PubMed
-
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. - PubMed
-
- Kyriakis JM, Banerjee P, Nikolakaki E, Dai T, Rubie EA, Ahmad MF, Avruch J, Woodgett JR. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. - PubMed
-
- Lee JC, Laydon JT, McDonnell PC, Gallagher TF, Kumar S, Green D, McNulty D, Blumenthal MJ, Heys JR, Landvatter SW, et al. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994 Dec 22–29;372:739–746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

