Natively unstructured loops differ from other loops
- PMID: 17658943
- PMCID: PMC1924875
- DOI: 10.1371/journal.pcbi.0030140
Natively unstructured loops differ from other loops
Abstract
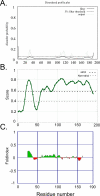
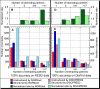
Natively unstructured or disordered protein regions may increase the functional complexity of an organism; they are particularly abundant in eukaryotes and often evade structure determination. Many computational methods predict unstructured regions by training on outliers in otherwise well-ordered structures. Here, we introduce an approach that uses a neural network in a very different and novel way. We hypothesize that very long contiguous segments with nonregular secondary structure (NORS regions) differ significantly from regular, well-structured loops, and that a method detecting such features could predict natively unstructured regions. Training our new method, NORSnet, on predicted information rather than on experimental data yielded three major advantages: it removed the overlap between testing and training, it systematically covered entire proteomes, and it explicitly focused on one particular aspect of unstructured regions with a simple structural interpretation, namely that they are loops. Our hypothesis was correct: well-structured and unstructured loops differ so substantially that NORSnet succeeded in their distinction. Benchmarks on previously used and new experimental data of unstructured regions revealed that NORSnet performed very well. Although it was not the best single prediction method, NORSnet was sufficiently accurate to flag unstructured regions in proteins that were previously not annotated. In one application, NORSnet revealed previously undetected unstructured regions in putative targets for structural genomics and may thereby contribute to increasing structural coverage of large eukaryotic families. NORSnet found unstructured regions more often in domain boundaries than expected at random. In another application, we estimated that 50%-70% of all worm proteins observed to have more than seven protein-protein interaction partners have unstructured regions. The comparative analysis between NORSnet and DISOPRED2 suggested that long unstructured loops are a major part of unstructured regions in molecular networks.
Conflict of interest statement
Figures



References
-
- Lesk AM. Introduction to protein architecture: The structural biology of proteins. Oxford: Oxford University Press; 2004. 347
-
- Brändén C, Tooze J. Introduction to protein structure. New York: Garland; 1991. 302
-
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. - PubMed
-
- Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. - PubMed
-
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

