Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions
- PMID: 17658949
- PMCID: PMC1924870
- DOI: 10.1371/journal.ppat.0030103
Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions
Abstract
Hepatitis C virus (HCV) infection is associated with chronic liver disease and currently affects about 3% of the world population. Although much has been learned about the function of individual viral proteins, the role of the HCV p7 protein in virus replication is not known. Recent data, however, suggest that it forms ion channels that may be targeted by antiviral compounds. Moreover, this protein was shown to be essential for infectivity in chimpanzee. Employing the novel HCV infection system and using a genetic approach to investigate the function of p7 in the viral replication cycle, we find that this protein is essential for efficient assembly and release of infectious virions across divergent virus strains. We show that p7 promotes virus particle production in a genotype-specific manner most likely due to interactions with other viral factors. Virus entry, on the other hand, is largely independent of p7, as the specific infectivity of released virions with a defect in p7 was not affected. Together, these observations indicate that p7 is primarily involved in the late phase of the HCV replication cycle. Finally, we note that p7 variants from different isolates deviate substantially in their capacity to promote virus production, suggesting that p7 is an important virulence factor that may modulate fitness and in turn virus persistence and pathogenesis.
Conflict of interest statement
Figures

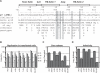


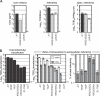

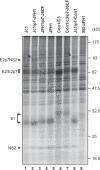
References
-
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42:962–973. - PubMed
-
- Hoofnagle JH. Course and outcome of hepatitis C. Hepatology. 2002;36:S21–S29. - PubMed
-
- Bartenschlager R, Frese M, Pietschmann T. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res. 2004;63:71–180. - PubMed
-
- Penin F, Dubuisson J, Rey FA, Moradpour D, Pawlotsky JM. Structural biology of hepatitis C virus. Hepatology. 2004;39:5–19. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

