Applying negative rule mining to improve genome annotation
- PMID: 17659089
- PMCID: PMC1940032
- DOI: 10.1186/1471-2105-8-261
Applying negative rule mining to improve genome annotation
Abstract
Background: Unsupervised annotation of proteins by software pipelines suffers from very high error rates. Spurious functional assignments are usually caused by unwarranted homology-based transfer of information from existing database entries to the new target sequences. We have previously demonstrated that data mining in large sequence annotation databanks can help identify annotation items that are strongly associated with each other, and that exceptions from strong positive association rules often point to potential annotation errors. Here we investigate the applicability of negative association rule mining to revealing erroneously assigned annotation items.
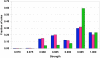
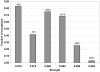
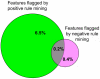
Results: Almost all exceptions from strong negative association rules are connected to at least one wrong attribute in the feature combination making up the rule. The fraction of annotation features flagged by this approach as suspicious is strongly enriched in errors and constitutes about 0.6% of the whole body of the similarity-transferred annotation in the PEDANT genome database. Positive rule mining does not identify two thirds of these errors. The approach based on exceptions from negative rules is much more specific than positive rule mining, but its coverage is significantly lower.
Conclusion: Mining of both negative and positive association rules is a potent tool for finding significant trends in protein annotation and flagging doubtful features for further inspection.
Figures
References
-
- Galperin MY, Koonin EV. Sources of systematic error in functional annotation of genomes: domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

