Cofactor of BRCA1 modulates androgen-dependent transcription and alternative splicing
- PMID: 17659869
- PMCID: PMC2701476
- DOI: 10.1016/j.jsbmb.2007.05.031
Cofactor of BRCA1 modulates androgen-dependent transcription and alternative splicing
Abstract
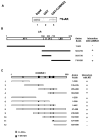
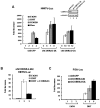
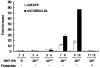
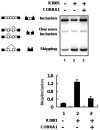
Transcriptional activity of nuclear receptors (NRs) is influenced by a large number of coregulators that exert their actions predominantly at the transcription initiation step. Unlike most well-characterized NR coregulators, cofactor of BRCA1 (COBRA1), a subunit of the negative elongation factor (NELF), binds to estrogen receptor alpha (ERalpha) and modulates estrogen-dependent transcription by impeding the movement of RNA polymerase II (RNAPII) during the transcription elongation stage. Here we show that, in addition to ERalpha, COBRA1 also displays various degrees of affinity for several other NRs. In particular, COBRA1 binds strongly to androgen receptor (AR) via its ligand-binding domain (LBD). Small hairpin RNA (shRNA)-mediated reduction of endogenous COBRA1 enhances androgen-mediated transcription. The effect of COBRA1 knockdown can be rescued by a silent mutant COBRA1 that is refractory to the shRNA action. Using a reporter assay for alternative splicing, we also provide evidence for a role of COBRA1 in influencing the exon skipping/inclusion of nascent transcripts produced from an androgen-dependent promoter. These findings suggest that COBRA1 may coordinate multiple steps in ligand-dependent gene expression, which in turn ensures both the quantity and quality of hormone-stimulated gene products.
Figures

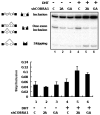
References
-
- Nilsson S, Makela S, Treuter E, Tujague M, Thomsen J, Andersson G, Enmark E, Pttersson K, Warner M, Gustafsson JA. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–1565. - PubMed
-
- Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes & Dev. 2000;14:121–141. - PubMed
-
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. - PubMed
-
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000;407:471–475. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

