Production of selenomethionyl-derivatized proteins in baculovirus-infected insect cells
- PMID: 17660253
- PMCID: PMC2206972
- DOI: 10.1110/ps.072931407
Production of selenomethionyl-derivatized proteins in baculovirus-infected insect cells
Abstract
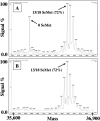
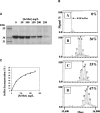
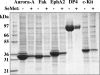
A protocol is described for the production of both intracellularly expressed and secreted selenomethionyl-derivatized recombinant proteins in baculovirus-infected insect cells. The method results in the production of recombinant soluble proteins with an SeMet occupancy of approximately 75% and with a recovery of approximately 20% that of native protein expression. The method is independent of the percentage methionine content of the protein and is reliable and consistent. Similar results are obtained using either Spodoptera frugiperda Sf9 or Trichoplusia ni High Five insect cells as the expression host, and when cultures are grown in either shake flasks or in Wave BioReactors.
Figures

Similar articles
-
A practical method for efficient and optimal production of Seleno-methionine-labeled recombinant protein complexes in the insect cells.Protein Sci. 2019 Apr;28(4):808-822. doi: 10.1002/pro.3575. Epub 2019 Feb 4. Protein Sci. 2019. PMID: 30663186 Free PMC article.
-
Production of recombinant adeno-associated viral vectors using a baculovirus/insect cell suspension culture system: from shake flasks to a 20-L bioreactor.Biotechnol Prog. 2005 Jan-Feb;21(1):154-60. doi: 10.1021/bp049802e. Biotechnol Prog. 2005. PMID: 15903253
-
The role of host cell physiology in the productivity of the baculovirus-insect cell system: Fluxome analysis of Trichoplusia ni and Spodoptera frugiperda cell lines.Biotechnol Bioeng. 2017 Mar;114(3):674-684. doi: 10.1002/bit.26089. Epub 2016 Sep 26. Biotechnol Bioeng. 2017. PMID: 27568545
-
Protein production using the baculovirus-insect cell expression system.Biotechnol Prog. 2014 Jan-Feb;30(1):1-18. doi: 10.1002/btpr.1842. Biotechnol Prog. 2014. PMID: 24265112 Review.
-
Baculovirus--insect cell interactions.Cytotechnology. 1996;20(1-3):73-93. doi: 10.1007/BF00350390. Cytotechnology. 1996. PMID: 8987578 Review.
Cited by
-
Mutation of high-affinity methionine permease contributes to selenomethionyl protein production in Saccharomyces cerevisiae.Appl Environ Microbiol. 2010 Oct;76(19):6351-9. doi: 10.1128/AEM.01026-10. Epub 2010 Aug 6. Appl Environ Microbiol. 2010. PMID: 20693451 Free PMC article.
-
Structure of an APC3-APC16 complex: insights into assembly of the anaphase-promoting complex/cyclosome.J Mol Biol. 2015 Apr 24;427(8):1748-64. doi: 10.1016/j.jmb.2014.11.020. Epub 2014 Dec 6. J Mol Biol. 2015. PMID: 25490258 Free PMC article.
-
Equi-MOI ratio for rapid baculovirus-mediated multiprotein co-expression in insect cells integrating selenomethionine for structural studies.FEBS Open Bio. 2025 Apr;15(4):563-572. doi: 10.1002/2211-5463.70025. Epub 2025 Mar 18. FEBS Open Bio. 2025. PMID: 40103323 Free PMC article.
-
Strategies for manufacturing recombinant adeno-associated virus vectors for gene therapy applications exploiting baculovirus technology.Brief Funct Genomic Proteomic. 2008 Jul;7(4):303-11. doi: 10.1093/bfgp/eln034. Epub 2008 Jul 16. Brief Funct Genomic Proteomic. 2008. PMID: 18632744 Free PMC article. Review.
-
A practical method for efficient and optimal production of Seleno-methionine-labeled recombinant protein complexes in the insect cells.Protein Sci. 2019 Apr;28(4):808-822. doi: 10.1002/pro.3575. Epub 2019 Feb 4. Protein Sci. 2019. PMID: 30663186 Free PMC article.
References
-
- Aertgerts K., Ye, S., Tennant, M.G., Kraus, M., Rogers, J., Sang, B.-C., Skene, R.J., Webb, D.R., and Prasad, G.S. 2004. Crystal structure of human dipeptidyl peptidase IV in complex with a decapeptide reveals details on substrate specificity and tetrahedral intermediate formation. Protein Sci. 13 412–421. - PMC - PubMed
-
- Aricescu A.R., Lu, W., and Jones, E.Y. 2006a. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D62 1243–1250. - PubMed
-
- Bellizzi J.J., Widom, J., Kemp, C.W., and Clardy, J. 1999. Producing selenomethionine-labeled proteins with a baculovirus expression vector system. Structure 7 R263–R267. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

