Probing the local conformational change of alpha1-antitrypsin
- PMID: 17660256
- PMCID: PMC2206966
- DOI: 10.1110/ps.072911607
Probing the local conformational change of alpha1-antitrypsin
Abstract
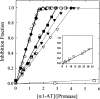
The native form of serpins (serine protease inhibitors) is a metastable conformation, which converts into a more stable form upon complex formation with a target protease. It has been suggested that movement of helix-F (hF) and the following loop connecting to strand 3 of beta-sheet A (thFs3A) is critical for such conformational change. Despite many speculations inferred from analysis of the serpin structure itself, direct experimental evidence for the mobilization of hF/thFs3A during the inhibition process is lacking. To probe the mechanistic role of hF and thFs3A during protease inhibition, a disulfide bond was engineered in alpha(1)-antitrypsin, which would lock the displacement of thFs3A from beta-sheet A. We measured the inhibitory activity of each disulfide-locked mutant and its heat stability against loop-sheet polymerization. Presence of a disulfide between thFs3A and s5A but not between thFs3A and s3A caused loss of the inhibitory activity, suggesting that displacement of hF/thFs3A from strand 5A but not from strand 3A is required during the inhibition process. While showing little influence on the inhibitory activity, the disulfide between thFs3A and s3A retarded loop-sheet polymerization significantly. This successful protein engineering of alpha(1)-antitrypsin is expected to be of value in clinical applications. Based on our current studies, we propose that the reactive-site loop of a serpin glides through between s5A and thFs3A for the full insertion into beta-sheet A while a substantial portion of the interactions between hF and s3A is kept intact.
Figures




Similar articles
-
Dimers initiate and propagate serine protease inhibitor polymerisation.J Mol Biol. 2008 Jan 4;375(1):36-42. doi: 10.1016/j.jmb.2007.10.055. Epub 2007 Oct 25. J Mol Biol. 2008. PMID: 18005992
-
Viscous drag as the source of active site perturbation during protease translocation: insights into how inhibitory processes are controlled by serpin metastability.J Mol Biol. 2006 Jun 2;359(2):378-89. doi: 10.1016/j.jmb.2006.03.045. Epub 2006 Apr 3. J Mol Biol. 2006. PMID: 16626735
-
Role of Lys335 in the metastability and function of inhibitory serpins.Protein Sci. 2000 May;9(5):934-41. doi: 10.1110/ps.9.5.934. Protein Sci. 2000. PMID: 10850803 Free PMC article.
-
Wild-type alpha 1-antitrypsin is in the canonical inhibitory conformation.J Mol Biol. 1998 Jan 23;275(3):419-25. doi: 10.1006/jmbi.1997.1458. J Mol Biol. 1998. PMID: 9466920 Review.
-
Alpha1-antitrypsin polymerization and the serpinopathies: pathobiology and prospects for therapy.J Clin Invest. 2002 Dec;110(11):1585-90. doi: 10.1172/JCI16782. J Clin Invest. 2002. PMID: 12464660 Free PMC article. Review. No abstract available.
Cited by
-
An antibody that prevents serpin polymerisation acts by inducing a novel allosteric behaviour.Biochem J. 2016 Oct 1;473(19):3269-90. doi: 10.1042/BCJ20160159. Epub 2016 Jul 12. Biochem J. 2016. PMID: 27407165 Free PMC article.
-
A hydrophobic patch surrounding Trp154 in human neuroserpin controls the helix F dynamics with implications in inhibition and aggregation.Sci Rep. 2017 Feb 23;7:42987. doi: 10.1038/srep42987. Sci Rep. 2017. PMID: 28230174 Free PMC article.
-
Local and global effects of a cavity filling mutation in a metastable serpin.Biochemistry. 2009 Sep 1;48(34):8233-40. doi: 10.1021/bi900342d. Biochemistry. 2009. PMID: 19624115 Free PMC article.
-
Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain.Chem Biol Drug Des. 2012 Nov;80(5):706-16. doi: 10.1111/cbdd.12010. Epub 2012 Aug 31. Chem Biol Drug Des. 2012. PMID: 22846652 Free PMC article.
-
Probing the folding pathway of a consensus serpin using single tryptophan mutants.Sci Rep. 2018 Feb 1;8(1):2121. doi: 10.1038/s41598-018-19567-9. Sci Rep. 2018. PMID: 29391487 Free PMC article.
References
-
- Abusriwil H. and Stockley, R.A. 2006. α1-Antitrypsin replacement therapy: Current status. Curr. Opin. Pulm. Med. 12 125–131. - PubMed
-
- Aulak K.S., Eldering, E., Hack, C.E., Lubbers, Y.P., Harrison, R.A., Mast, A., Cicardi, M., and Davis 3rd, A.E. 1993. A hinge region mutation in C1-inhibitor (A436T) results in nonsubstrate-like behavior and in polymerization of the molecule. J. Biol. Chem. 268 18088–18094. - PubMed
-
- Baumann U., Huber, R., Bode, W., Grosse, D., Lesjak, M., and Laurell, C.B. 1991. Crystal structure of cleaved human α1-antichymotrypsin at 2.7 Å resolution and its comparison with other serpins. J. Mol. Biol. 218 595–606. - PubMed
-
- Beatty K., Bieth, J., and Travis, J. 1980. Kinetics of association of serine proteinases with native and oxidized α1-proteinase inhibitor and α1-antichymotrypsin. J. Biol. Chem. 255 3931–3934. - PubMed
-
- Bijnens A.P., Gils, A., Knockaert, I., Stassen, J.M., and Declerck, P.J. 2000. Importance of the hinge region between α1-helix F and the main part of serpins, based upon identification of the epitope of plasminogen activator inhibitor type 1 neutralizing antibodies. J. Biol. Chem. 275 6375–6380. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

