Genetic analysis of the E site during RF2 programmed frameshifting
- PMID: 17660276
- PMCID: PMC1950767
- DOI: 10.1261/rna.638707
Genetic analysis of the E site during RF2 programmed frameshifting
Erratum in
- RNA. 2007 Nov;13(11):2051
Abstract
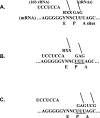
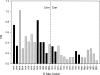
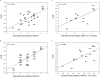
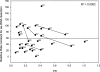
The roles of the ribosomal E site are not fully understood. Prior evidence suggests that deacyl-tRNA in the E site can prevent frameshifting. We hypothesized that if the E-site codon must dissociate from its tRNA to allow for frameshifting, then weak codon:anticodon duplexes should allow for greater frameshifting than stronger duplexes. Using the well-characterized Escherichia coli RF2 (prfB) programmed frameshift to study frameshifting, we mutagenized the E-site triplet to all Unn and Cnn codons. Those variants should represent a very wide range of duplex stability. Duplex stability was estimated using two different methods. Frameshifting is inversely correlated with stability, as estimated by either method. These findings indicate that pairing between the deacyl-tRNA and the E-site codon opposes frameshifting. We discuss the implications of these findings on frame maintenance and on the RF2 programmed frameshift mechanism.
Figures

Similar articles
-
Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2.Cell. 2004 Jul 9;118(1):45-55. doi: 10.1016/j.cell.2004.06.012. Cell. 2004. PMID: 15242643
-
Analysis of effects of tRNA:message stability on frameshift frequency at the Escherichia coli RF2 programmed frameshift site.Nucleic Acids Res. 1993 Apr 25;21(8):1837-43. doi: 10.1093/nar/21.8.1837. Nucleic Acids Res. 1993. PMID: 8493101 Free PMC article.
-
mRNA-Mediated Duplexes Play Dual Roles in the Regulation of Bidirectional Ribosomal Frameshifting.Int J Mol Sci. 2018 Dec 4;19(12):3867. doi: 10.3390/ijms19123867. Int J Mol Sci. 2018. PMID: 30518074 Free PMC article.
-
Ribosomal frameshifting, jumping and readthrough.Curr Opin Cell Biol. 1991 Dec;3(6):1051-5. doi: 10.1016/0955-0674(91)90128-l. Curr Opin Cell Biol. 1991. PMID: 1814364 Free PMC article. Review.
-
How translational accuracy influences reading frame maintenance.EMBO J. 1999 Mar 15;18(6):1427-34. doi: 10.1093/emboj/18.6.1427. EMBO J. 1999. PMID: 10075915 Free PMC article. Review.
Cited by
-
Codon-Anticodon Recognition in the Bacillus subtilis glyQS T Box Riboswitch: RNA-DEPENDENT CODON SELECTION OUTSIDE THE RIBOSOME.J Biol Chem. 2015 Sep 18;290(38):23336-47. doi: 10.1074/jbc.M115.673236. Epub 2015 Jul 30. J Biol Chem. 2015. PMID: 26229106 Free PMC article.
-
Ribosome collisions alter frameshifting at translational reprogramming motifs in bacterial mRNAs.Proc Natl Acad Sci U S A. 2019 Oct 22;116(43):21769-21779. doi: 10.1073/pnas.1910613116. Epub 2019 Oct 7. Proc Natl Acad Sci U S A. 2019. PMID: 31591196 Free PMC article.
-
Shine-Dalgarno interaction prevents incorporation of noncognate amino acids at the codon following the AUG.Proc Natl Acad Sci U S A. 2008 Aug 5;105(31):10715-20. doi: 10.1073/pnas.0801974105. Epub 2008 Jul 30. Proc Natl Acad Sci U S A. 2008. PMID: 18667704 Free PMC article.
-
Structured mRNA induces the ribosome into a hyper-rotated state.EMBO Rep. 2014 Feb;15(2):185-90. doi: 10.1002/embr.201337762. Epub 2014 Jan 8. EMBO Rep. 2014. PMID: 24401932 Free PMC article.
-
Frameshifting at collided ribosomes is modulated by elongation factor eEF3 and by integrated stress response regulators Gcn1 and Gcn20.RNA. 2022 Mar;28(3):320-339. doi: 10.1261/rna.078964.121. Epub 2021 Dec 16. RNA. 2022. PMID: 34916334 Free PMC article.
References
-
- Agris, P.F., Vendeix, F.A.P., Graham, W.D. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007;366:1–13. - PubMed
-
- Björk, G.R. Biosynthesis and function of modified nucleosides. In: Söll D., RajBhandary U., editors. tRNA: Structure, biosynthesis and function. ASM Press; Washington, DC: 1995. pp. 165–205.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
